-
煤炭一直以来是我国主要的能源物质[1],而洗选是实现煤炭清洁化利用的源头技术[2]. 目前选煤厂仍以湿法选煤为主,伴随产生大量的煤泥水亟需澄清处理以实现水的循环[3]. 阴离子型聚丙烯酰胺(HPAM)是由单体丙烯酰胺(AM)和丙烯酸(AA)在碱性条件下共聚而成的一种线性水溶性聚合物,分子量在300万—2000万区间[4 − 5]. HPAM作为选煤絮凝剂,其常用于煤泥水的絮凝沉降[6 − 7]. 随着HPAM的广泛使用,伴随产生大量的含聚污水,增加了处理难度及成本[8]. 煤泥水中的悬浮颗粒通过吸附到HPAM而实现桥连,形成较大絮团以促进分离[9]. HPAM本身被认为是无毒的[10],但已证明其暴露在紫外线或光、热时会缓慢发生降解,释放出少量有毒的AM,该物质将会损伤人和动物的神经系统[11]. 此外,HPAM高分子量的碳链具有生物抗性,较难生物降解,导致其在选煤厂周边环境中大量累积,这将对生态环境构成潜在威胁[12]. 所以,有必要将HPAM降解转化成无毒性的小分子物质.
HPAM降解处理方法包括物理降解[13]、化学降解[14]、生物降解[15]及它们的结合等,而低成本、无二次污染、环境友好的生物降解转化常用于处理HPAM[16]. HPAM降解微生物通常从长期使用HPAM的环境中分离出来的,如HPAM处理的油田采出水、油砂尾矿、土壤、活性污泥等[17]. 生物降解本质是酶催化反应,同时在降解HPAM发酵液中检测到关键酶活性,如酰胺酶、脱氢酶、漆酶等. HPAM的生物降解开始于酰胺酶催化HPAM脱氨成氨和聚丙烯酸(PAA),同时氨可释放出来供微生物生长所需的氮源. HPAM的碳骨架PAA比其酰胺部分更难被生物降解[18],然而仍存在一些微生物是可以在HPAM和PAA的环境下生长[19]. HPAM经酰胺酶脱氨基反应后,残留的碳骨架PAA在加氧酶或氧化酶的作用下其碳主链被氧化断裂,类似于脂肪酸的α-C氧化[20]. Song等[21]通过实验发现由细菌分泌漆酶的活性不受HPAM浓度影响,这可能是由于该酶不仅能提高污染物的氧化速率,还能扩大氧化底物范围. 在好氧环境下漆酶可以从HPAM分子上夺得4个e−,将O2还原成H2O. 国内外学者筛选高效菌株并优化工艺尝试对废水中HPAM的降解处理已取得很大的进步,然而菌株的筛选及工艺优化任务繁重且缺乏科学依据,迄今为止关于生物酶学分析很大程度上是未知的,仅提出了一些假设的降解途径,确切的降解机理还不清楚. 同时,由于酶解是在水溶液中进行,生物酶与HPAM的微观作用分子机制仍鲜为人知,水环境对HPAM与酶相互作用的影响鲜有报道. 近年来,研究人员主要通过试验研究微生物降解HPAM的规律,例如淀粉-碘化镉、GPC/SEC、FT-IR、HPLC、扫描电镜(SEM)、黏度等方法,然而这些常规实验解释酶降解机理仍很困难. 漆酶的三维晶体结构已经通过实验解析出来,这为进一步在分子水平上探究其与底物相互作用提供了可能性[22]. 分子模拟可以在分子或原子尺度上探究酶与底物的相互作用,可以深入了解酶降解的分子机制,这将有助于解决HPAM降解效果差的问题[23 − 24].
本文采用半柔性对接探究枯草芽孢杆菌漆酶(Lac)与HPAM或PAA的结合,从最佳对接构象中筛选到最适酶与底物复合物,然后对该最适复合物分别进行基于亲和力的虚拟突变以及不同温度下的分子动力学(MD)模拟. 该研究的数据有助于深入理解酶降解机理,以期对HPAM及其中间产物PAA进行彻底降解.
-
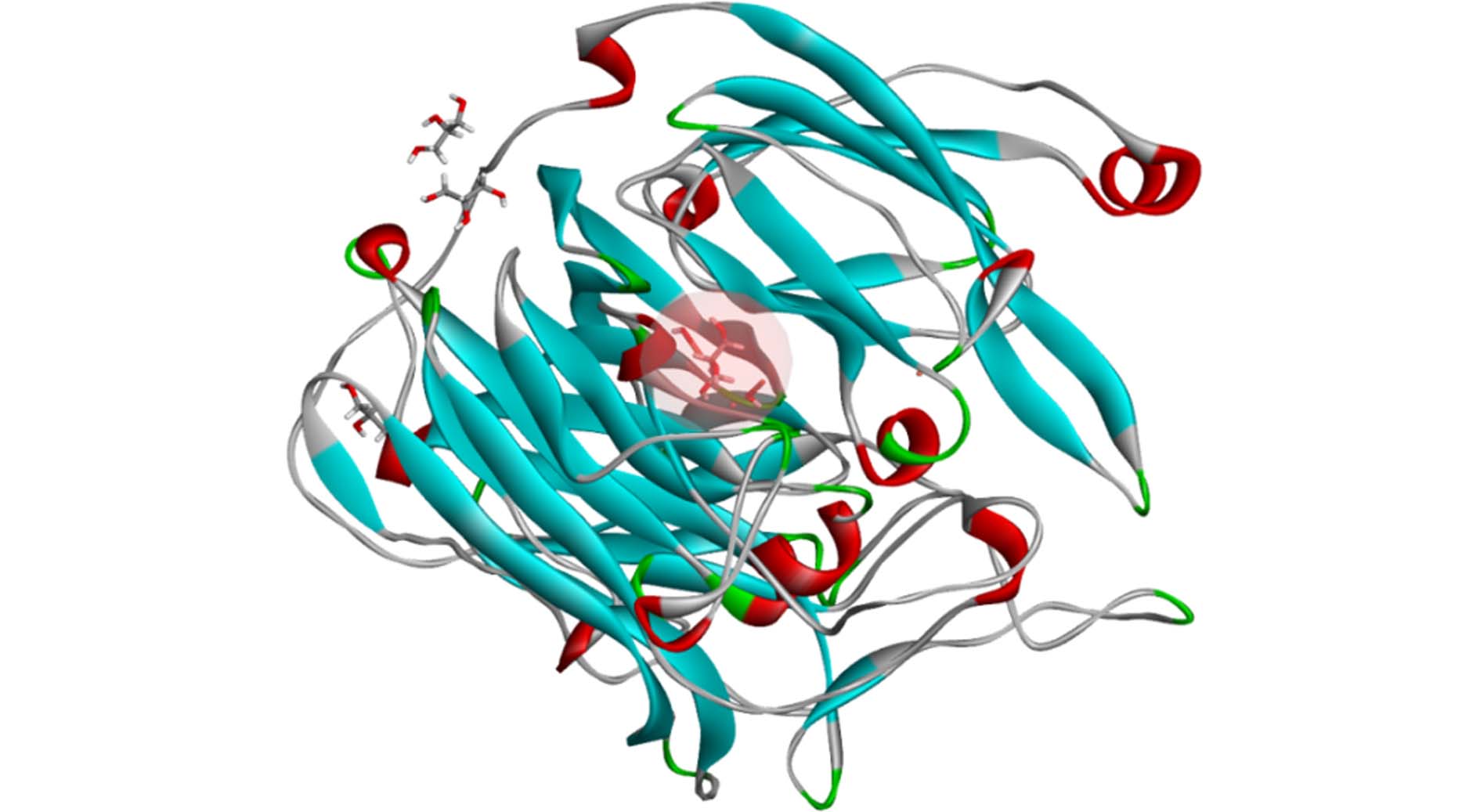
漆酶的三维晶体结构下载于蛋白质数据库(RCSB),相应PDB编号为1GSK,分辨率为0.170 nm,A链,该酶来源于枯草芽孢杆菌,简称Lac [25]. 采用Discovery Studio (DS) 2020的Prepare Protein工具对其进行去水加氢,修补缺失残基,如图1所示. 从图1可以看出,Lac三维晶体结构中含有4个原始配体丙三醇,透明红色球体显示丙三醇的位置离Lac的3个铜离子最近,而且已经证明漆酶的催化活性是与铜离子有关. 因此将透明红色球体的位置作为该酶对接的结合位点,其坐标为(X,
97.3796 ; Y,61.3005 ; Z, -8.42748 ; Radius, 5). 此外,对接前应删除所有原始配体. -
HPAM在弱碱性环境下其羧基侧链带负电. PAA实际上是HPAM的脱氨产物. HPAM和PAA皆是由重复的结构单元构成的线性聚合物,而且生物酶降解主要涉及官能团之间发生的反应,所以仅考虑酶与底物分子主链或侧链上的高反应活性官能团之间的相互作用即可.
本文选取具有代表性的结构单元为2—5的HPAM和PAA作为结构模型,它们的结构模型采用Materials Studio(MS) 2017软件的Build polymer工具绘制,两端采用甲基进行封端,同样的处理见文献[26]. 采用DS 2020的Prepare Ligands工具对所有配体进行预处理,以期获得正确的离子化状态下的三维结构,然后采用DS 2020的Minimize Ligands对它们在CHARMm力场下进行构型优化. 图2显示了几种不同分子链长度的HPAM和PAA模型物化学结构及其优化构型.
-
采用DS 2020的CDOCKER程序进行Lac与HPAM或PAA的分子对接模拟实验[27]. 该对接程序主要采用负总能打分(-CDOCKER_Energy score)和负相互作用能打分(-CDOCKER_Interaction _Energy score)两种能量打分评估对接结果. 其中,负总能打分值越大,说明酶同底物亲和力越大、越稳定[28]. 结果默认保留底物最优的前10种构象,选择-CDOCKER_ Energy score值最大的底物构象进行进一步分析.
-
底物与活性位点处残基的非键作用力距离一般取0.5 nm. 突变能是复合物突变前后总能量变化. 由对接筛选得到拥有-CDOCKER_ Energy score值最大的复合物,该复合物亲和力最大、最稳定. 根据酶促动力学理论推测出,此时底物是酶的最适底物. 为了进一步确定最适酶-底物复合物中的关键残基,采用DS 2020的Calculate Mutation Energy (Binding)程序对配体周围0.5 nm范围内残基进行丙氨酸(ALA)扫描,选择突变能大于0.5 kcal·mol−1的残基作为该酶结合底物的关键残基,然后对这些关键残基进行虚拟饱和突变,最终选择突变能小于−0.5 kcal·mol−1的残基作为理论设计高活性酶的突变位点.
-
MD可以用于探究温度对酶与底物随时间的动态变化影响,同时能够找出酶的最大波动残基区域. 酶与底物亲和力越大,Km值就越小,此时底物是酶的最适底物. 采用GROMACS软件包(版本
2021.5 )对最适酶与底物复合物进行298 K、303 K及308 K下的MD模拟[29],力场选为Amber03 force field [30]. 为使模拟过程更接近真实情况,体系被溶剂化放置在10 nm×10 nm×10 nm简单点电荷(SPC)水模型[31]周期性边界条件的立方盒子里,添加抗衡离子Na+和Cl−以中和体系. 然后,采用最速下降法进行能量最小化,并且在1个原子的恒压下进行了1 ns NPT MD使体系达到平衡. 最后,模拟体系进行了NPT MD 50 ns采样并且进行轨迹的数据分析. 采用gromacs tool gmx rmsd及gmx rmsf分别来计算酶骨架原子的RMSD和酶残基RMSF,同时采用DS 2020的Calculate Interaction Energy程序计算酶与底物相互作用能及残基对相互作用能的贡献. 用Visual Molecular Dynamics(VMD)软件对复合物的构象变化进行可视化分析及比较. MD模拟过程中范德华截断半径为1.4 nm,长程静电相互作用采用Particle Mesh Eward(PME)进行处理[32],步长设置为2.0 fs. 温度和压力分别采用Berendsen thermostat 和Berendsen barostat来维持,以上模拟均在Max-Flow平台下进行. -
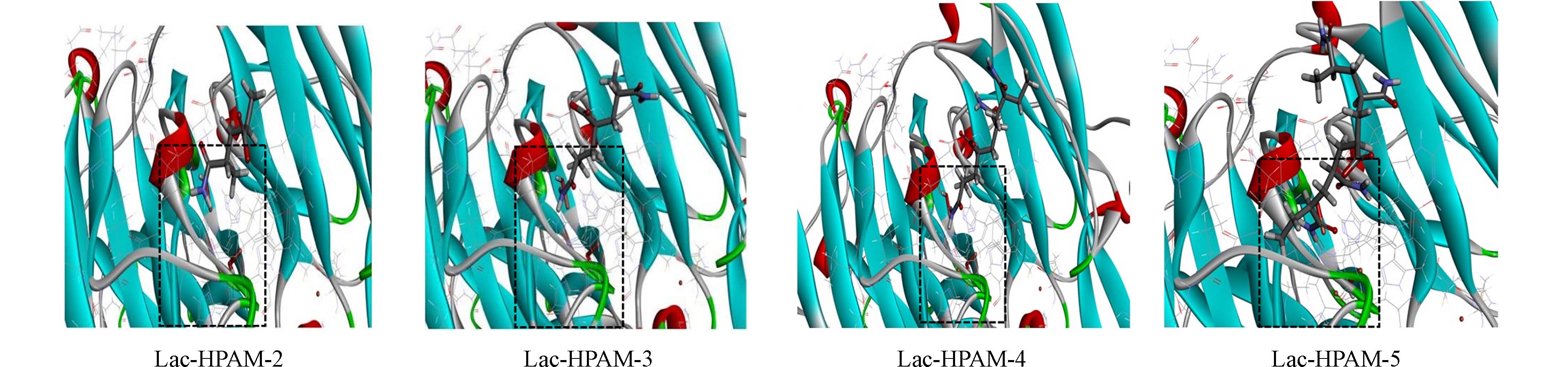
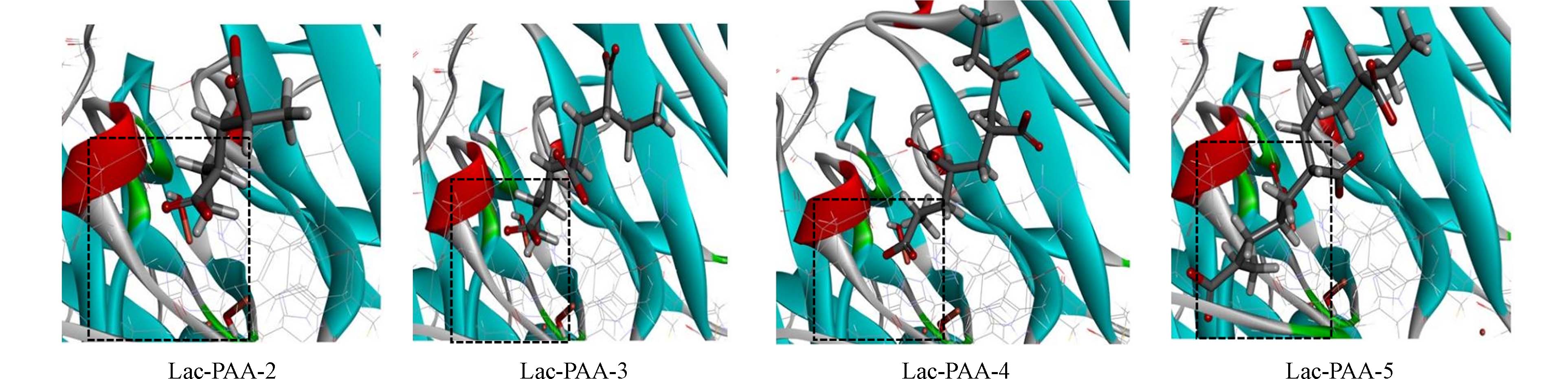
本研究中采用重复单元为2—5的HPAM和PAA的优化结构模型用于研究Lac与底物的相互作用. Lac与底物结合时亲和力越大、越稳定,越说明该酶降解底物的活性就越强. 从表1对接结果中的打分值可以看出,无论是基于 -CDOCKER_Energy score还是-CDOCKER_Interaction_Energy score指标来看,Lac-HPAM-3都拥有最高的打分,说明Lac对HPAM-3有最高的亲和力[33],此时HPAM-3是Lac的最适底物,而且它们相互作用最小,结合的最好. Lac对HPAM的能量打分皆随其碳链延长而先增加后减少,尤其该酶对HPAM-5的打分迅速降至最低. 这说明Lac对HPAM碳链长度有一定耐受性,倾向氧化降解HPAM的碳链,但HPAM-5很难被该酶降解,这也符合聚合物难于被酶解的规律.
当-CDOCKER_ Energy score和-CDOCKER_ Interaction_ Energy score呈现出不同的变化时,-CDOCKER_Energy score打分更值得信赖. 从表1中Lac与PAA结合时的-CDOCKER_Energy score值来看,可以发现Lac-PAA-2享有最高-CDOCKER_Energy score值(
24.8628 kcal·mol−1),但皆低于Lac-HPAM-3的打分(31.8442 kcal·mol−1)和Lac-HPAM-4的打分(28.8940 kcal·mol−1),说明PAA比HPAM更难被Lac降解. 实验表明,基于淀粉-碘化镉和TOC的结果的显著差异,细菌更倾向将HPAM的酰胺基水解成羧基作为氮源,但难以利用HPAM的碳链骨架作为碳源[18]. 即细菌很难利用其脱氨产物PAA作为营养物质. 无论基于 -CDOCKER_ Energy score还是-CDOCKER_Interaction_Energy score指标来看,Lac-PAA-5皆为负值,说明高分子量的PAA难于被Lac降解,同样也符合聚合物难于酶解的规律.图3显示了HPAM与Lac最佳的结合构象,Lac活性位点处的残基由线性模型显示. 从图3可以发现, HPAM-2、HPAM-3和HPAM-4的分子链舒展于活性位点附近,这种现象解释了Lac-HPAM-2、Lac-HPAM-3和Lac-HPAM-4能量打分皆为正,而且-CDOCKER_Energy score打分值在24.00—32.00 kcal·mol−1范围内. 这说明Lac可容纳较长HPAM碳链,倾向于降解一定长度寡聚物. 同时也观察到,只有HPAM-5偏离原来的对接位置,其它配体的对接位置均一致,这能很好地解释Lac-HPAM-5享有负的-CDOCKER_ Energy score值(−109.688 kcal·mol−1)的原因,进一步说明高分子量的HPAM很难被Lac降解.
图4显示了PAA与Lac最佳的结合构象,其中线性模型显示Lac活性位点处的残基. 从图4可见,PAA-2、PAA-3和PAA-4的分子链舒展于活性位点附近,这种现象解释了Lac-PAA-2、Lac-PAA-3 和 Lac-PAA-4 能量打分皆为正,而且-CDOCKER_Energy score打分值在17.00—25.00 kcal·mol−1范围,明显低于相应的Lac与HPAM结合. 这说明Lac可以容纳较长PAA碳链,但该酶更倾向于降解一定长度的HPAM. 同时也观察到,只有PAA-5偏离原来的对接位置,而其它配体对接位置一致,这能很好地解释为什么Lac-PAA-5享有负的-CDOCKER_Energy score(−152.526 kcal·mol−1)和-CDOCKER_ Interaction_Energy score(−
25.3122 kcal·mol−1),这说明高分子量PAA很难被Lac降解. -
(1)Lac与HPAM的结合
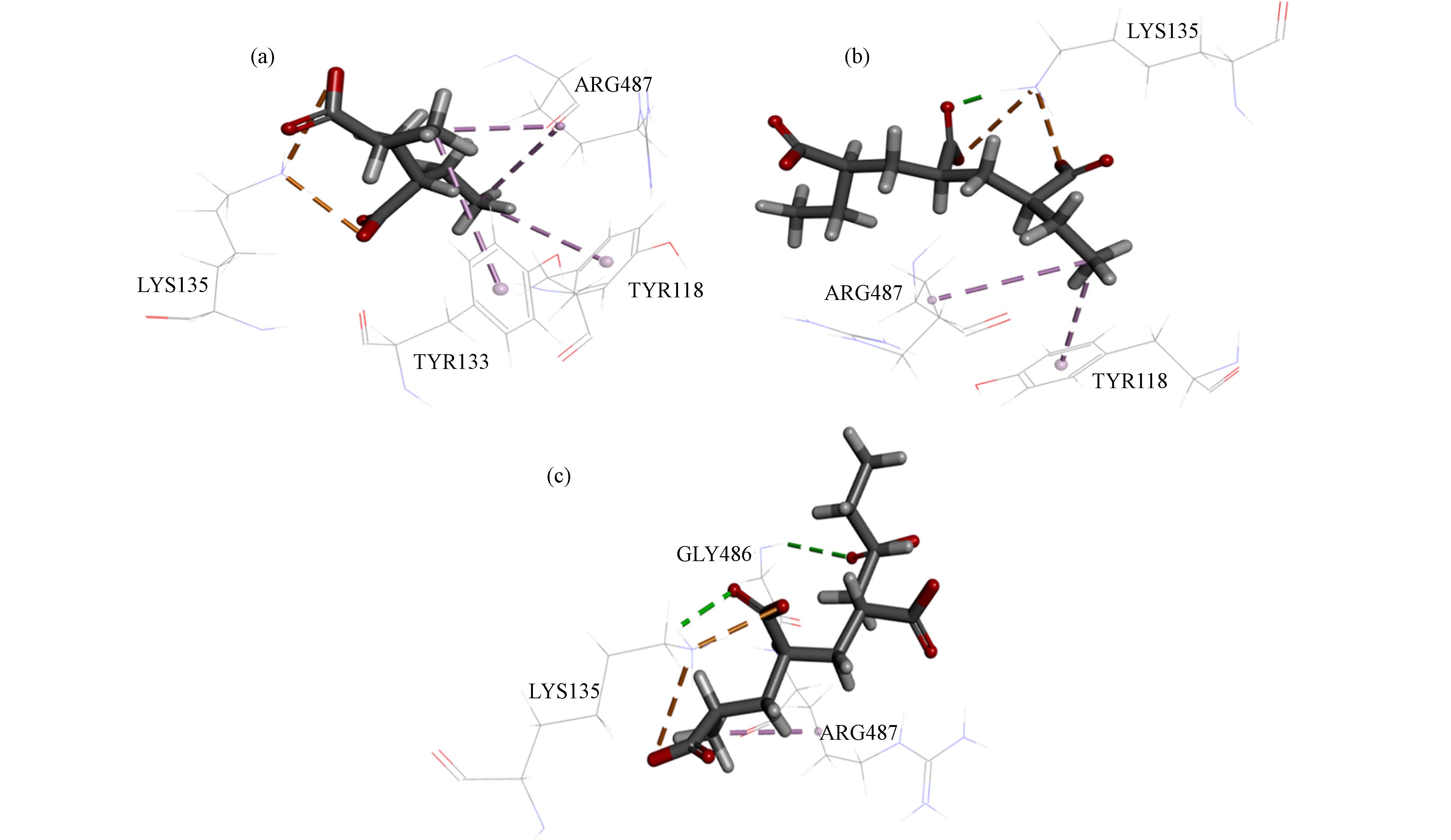
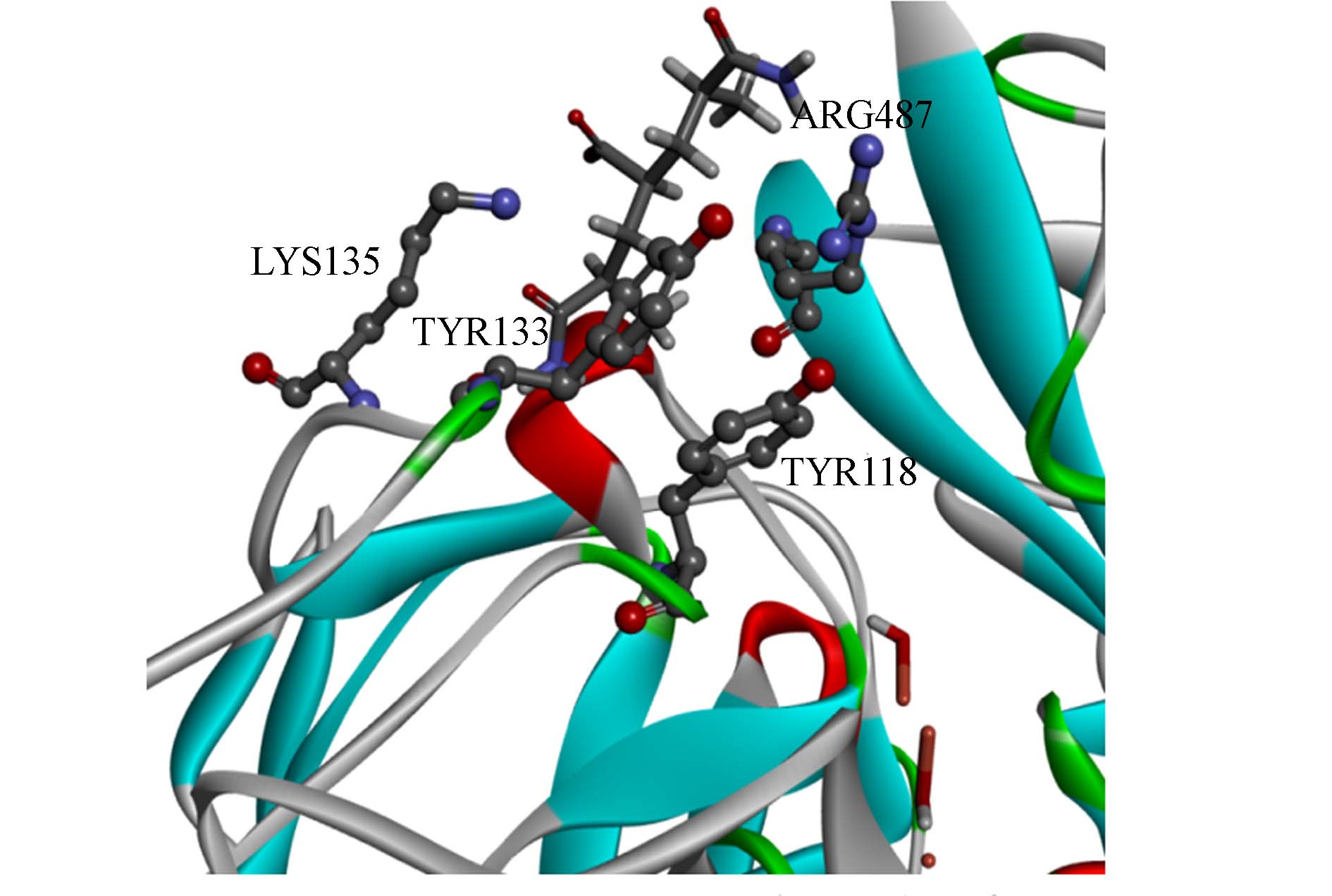
通过DS 2020将能量打分皆为正的Lac与HPAM复合物进行可视化,显示其相互作用(图5). 从图5可以观察到,HPAM-2与LYS135形成2个氢键,其与TYR133形成1个氢键,一共形成3个氢键. HPAM-2分子链两端与ARG487形成2个疏水键,其分子链一端分别与TYR118和TYR133各自形成1个疏水键,一共形成4个疏水键. HPAM-2-O−与LYS135-N+形成1个静电吸引;HPAM-3分别与TYR133和ASP507各自形成1个氢键,一共形成2个氢键. HPAM-3高分子链两端与ARG487形成2个疏水键. HPAM-3-O−与LYS135-NH+形成1个静电吸引,同时形成盐桥;HPAM-4分别与TYR133和LYS135各自形成1个氢键,一共形成2个氢键. HPAM-4高分子链一端与ARG487形成1个疏水键. HPAM-4-O−与LYS135-N+形成1个静电吸引.
通过对比发现, Lac-HPAM-3亲和力最大、最稳定的主要原因是HPAM-3与Lac之间形成强盐桥造成的,此时HPAM-3是Lac降解HPAM的最适底物. Lac与HPAM结合亲和力强度与其形成的氢键、疏水键和静电作用等数量及距离有关. 由于HPAM-2与Lac周围残基形成的疏水键较密集,而且疏水键数量最多,这将有利于Lac结合HPAM-2. 但HPAM-2与LYS135同时形成2个氢键,这将很大程度上不利于LYS135-NH+与HPAM-2-O−形成盐桥,只存在静电吸引. HPAM-4较长的分子链一端与ARG487形成一个较强的疏水作用,可以保证HPAM-4以一定的自由度结合在Lac的活性位点上;HPAM-3和HPAM-4与Lac周围的氨基酸残基形成2个氢键比较分散,这样有利于Lac结合HPAM. HPAM-4与LYS135形成1个氢键,它不利于LYS135-NH+与HPAM-4-O−形成盐桥,但其减弱程度低于HPAM-2,这可以很好地解释为什么Lac-HPAM-2结合亲和力最差、最不稳定. 通过上述分析,可以推测出亲和力大小的顺序为Lac-HPAM-3>Lac-HPAM-4>Lac-HPAM-2,这与CDOCKER对接程序获得的能量打分分析结果相一致.
(2)Lac与PAA的结合
通过DS 2020将能量打分皆为正的Lac与PAA复合物显示其相互作用细节(图6). 从图6可以观察到,PAA-2分子链两端与ARG487形成2个疏水键,其分子链一端分别与TYR118和TYR133各自形成1个疏水键,一共形成4个疏水键. PAA-2-O−与LYS135-N+形成2个静电吸引;PAA-3高分子链一端分别与ARG487和TYR118形成1个疏水键,一共形成2个疏水键. PAA-3-O−也与LYS135-N+形成2个静电吸引,同时LYS135与PAA-3形成1个氢键;PAA-4高分子链一端与ARG487形成1个疏水键. PAA-4-O−与LYS135-N+形成2个静电吸引,而LYS135与PAA-4形成1个氢键. PAA-4与GLY486形成1个氢键,一共形成2个氢键.
通过对比发现,PAA-2与Lac周围氨基酸残基形成的疏水作用数最多,而且密集,说明疏水作用是Lac-PAA-2最稳定、亲和力最大的主要原因,此时底物PAA-2是Lac降解PAA的最适底物. 另外,PAA-2-O−与LYS135-N+的两个静电吸引也可以较好地将其稳定在Lac的活性中心上. PAA-3和PAA-4都与LYS135形成1个氢键,这将会大大增加了它们与Lac的结合. 虽然Lac-PAA-3比Lac-PAA-4多了1个疏水作用,但Lac-PAA-4比Lac-PAA-3多了1个氢键作用. 所以,Lac-PAA-3和Lac-PAA-4的能量评分相差不多,进一步说明Lac可容纳一定链长度的PAA.
将上述分析的非键相互作用结果列于表2. 从表2可以观察到, TYR133参与Lac与HPAM中氢键的形成,而ARG487通过形成疏水、LYS135通过形成静电吸引或盐桥以及氢键参与所有复合物的稳定.
-
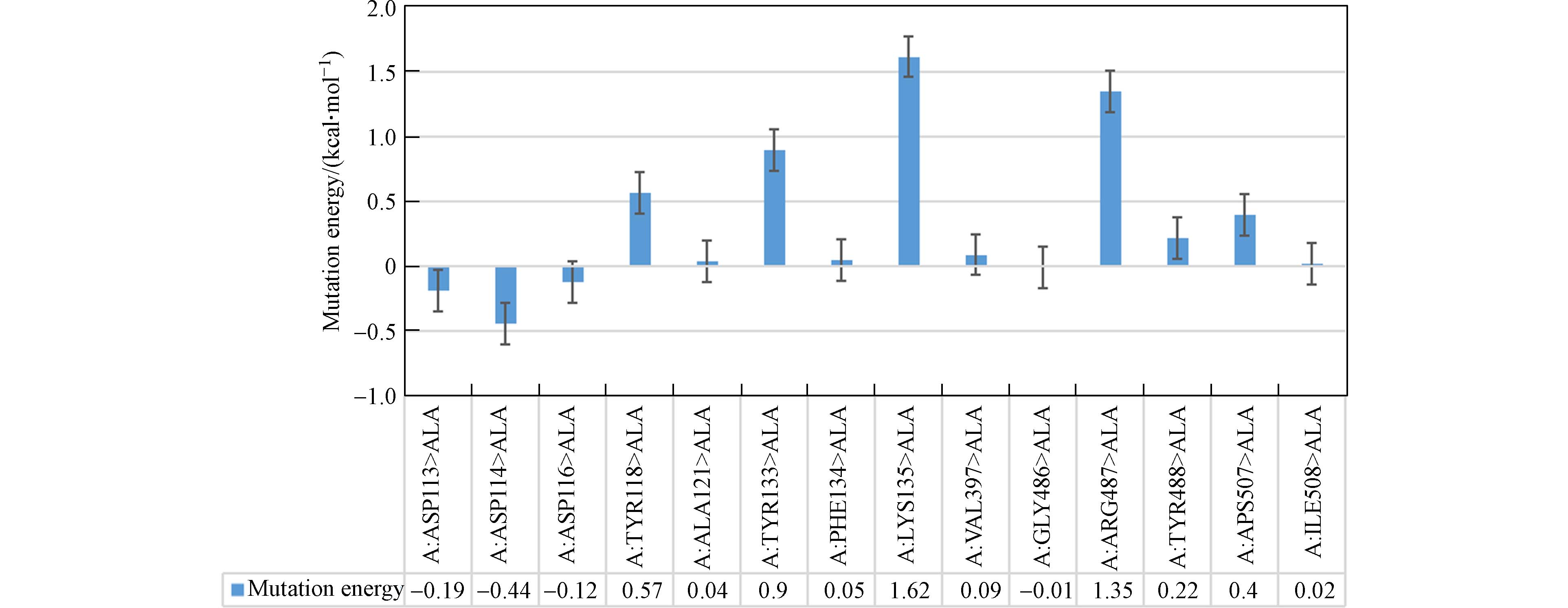
通过上述的分析得到Lac-HPAM-3的亲和力最大、最稳定,即HPAM-3是Lac的最适底物,所以采用DS 2020的Calculate Mutation Energy (Binding)模块对该复合物配体HPAM-3周围0.5 nm内的残基进行ALA扫描. 突变能作为唯一标准来评估单点突变对Lac同HPAM-3亲和力的影响. 图7显示了该复合物ALA扫描的所有突变能,而表3仅列出了其5个最高突变能和5个最低突变能. 从Lac-HPAM-3的ALA扫描结果可以发现,TYR118ALA、TYR133ALA、ARG487ALA和LYS135ALA等突变体的突变能都大于0.5 kcal·mol−1,效果为不稳定,说明这些突变可能会导致Lac与HPAM-3亲和力下降,由此可以判断出TYR118、TYR133、ARG487和LYS135等4种残基是Lac与HPAM-3结合的关键氨基酸残基. 其中,突变体LYS135ALA的突变能(1.62 kcal·mol−1)最大,说明LYS135对Lac与HPAM-3亲和力影响最大,即LYS135与HPAM-3形成的盐桥对Lac-HPAM-3亲和力影响最大,进一步说明盐桥作用是Lac-HPAM-3亲和力最大的主要原因. 而突变体ARG487ALA的突变能(1.35 kcal·mol−1)仅次于突变体LYS135ALA,说明ARG487参与的疏水作用对Lac与HPAM-3亲和力影响也较大. 图8显示了Lac结合HPAM-3时该酶关键残基的位置.
-
采用DS 2020的Calculate Mutation Energy (Binding)模块对Lac-HPAM-3的关键残基进行虚拟饱和突变. 即将TYR118、TYR133、ARG487和LYS135等关键残基分别突变为除ALA外其余的19种标准残基,虚拟饱和突变结果列于表4,而且仅列出了5个最高的突变能和5个最低的突变能. 当TYR118LYS、LYS135ASP、LYS135TYR、LYS135HIS和ARG487PRO时,突变能分别为2.42、2.50、2.52、3.13、4.64 kcal·mol−1,这些突变能有较大值,都大于2.40 kcal·mol−1,影响均为不稳定,说明这些突变会使Lac对HPAM-3的亲和力下降,相互作用减弱,最终导致Lac的活性降低. 其中有3种突变体是包含残基LYS135对亲和力的影响,说明LYS135参与的盐桥对稳定Lac-HPAM-3起关键作用. 此外,突变体ARG487PRO的突变能(4.64 kcal·mol−1)最大,说明ARG487参与的疏水作用也对稳定Lac-HPAM-3起关键作用.
-
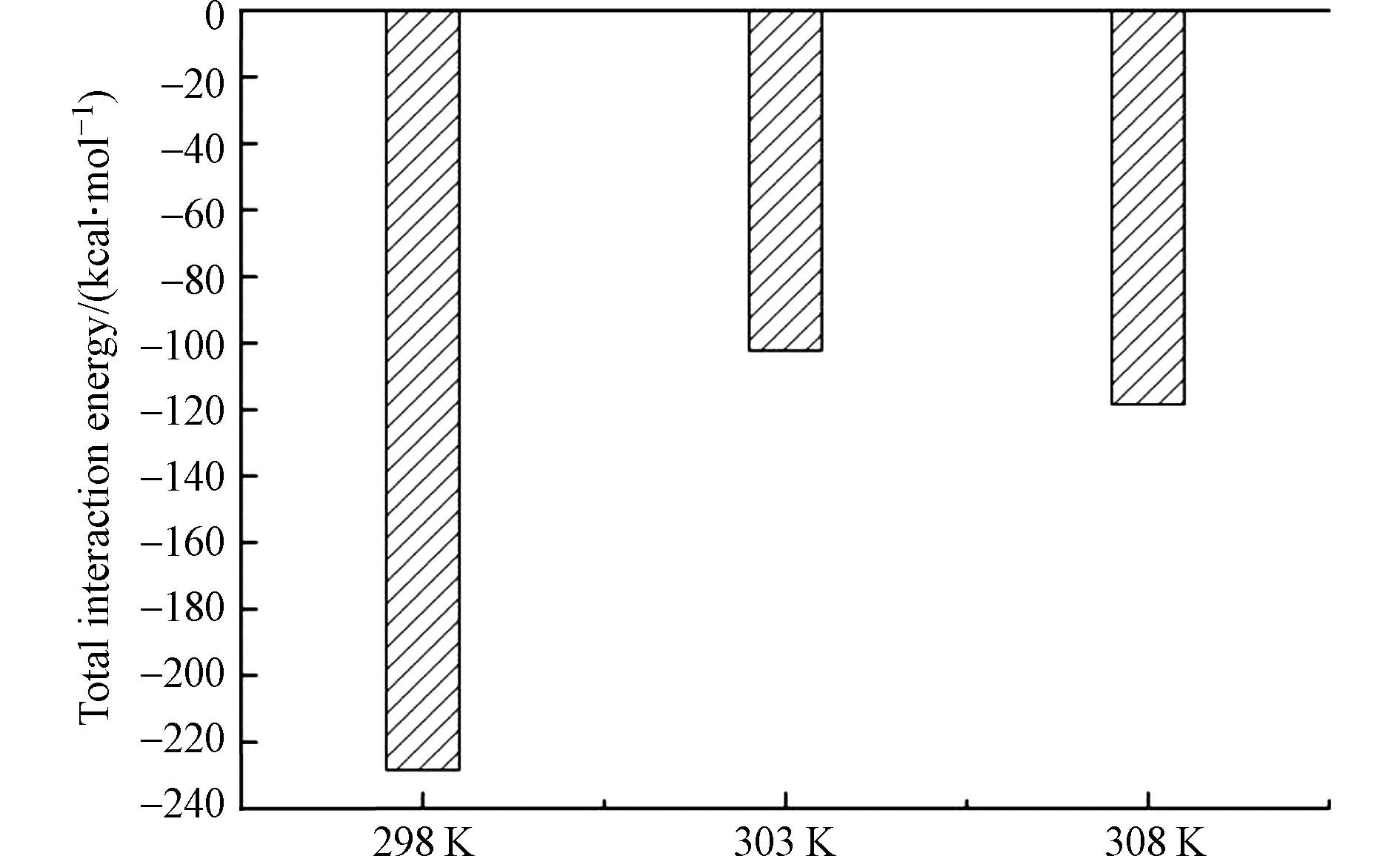
通过对接可以得到Lac的最适底物是HPAM-3,因此不同温度下它们的总相互作用能如图9所示. 从图9可以看到,298 K时Lac与HPAM-3的总相互作用能最小,说明Lac与HPAM-3在298 K时结合的最好.
-
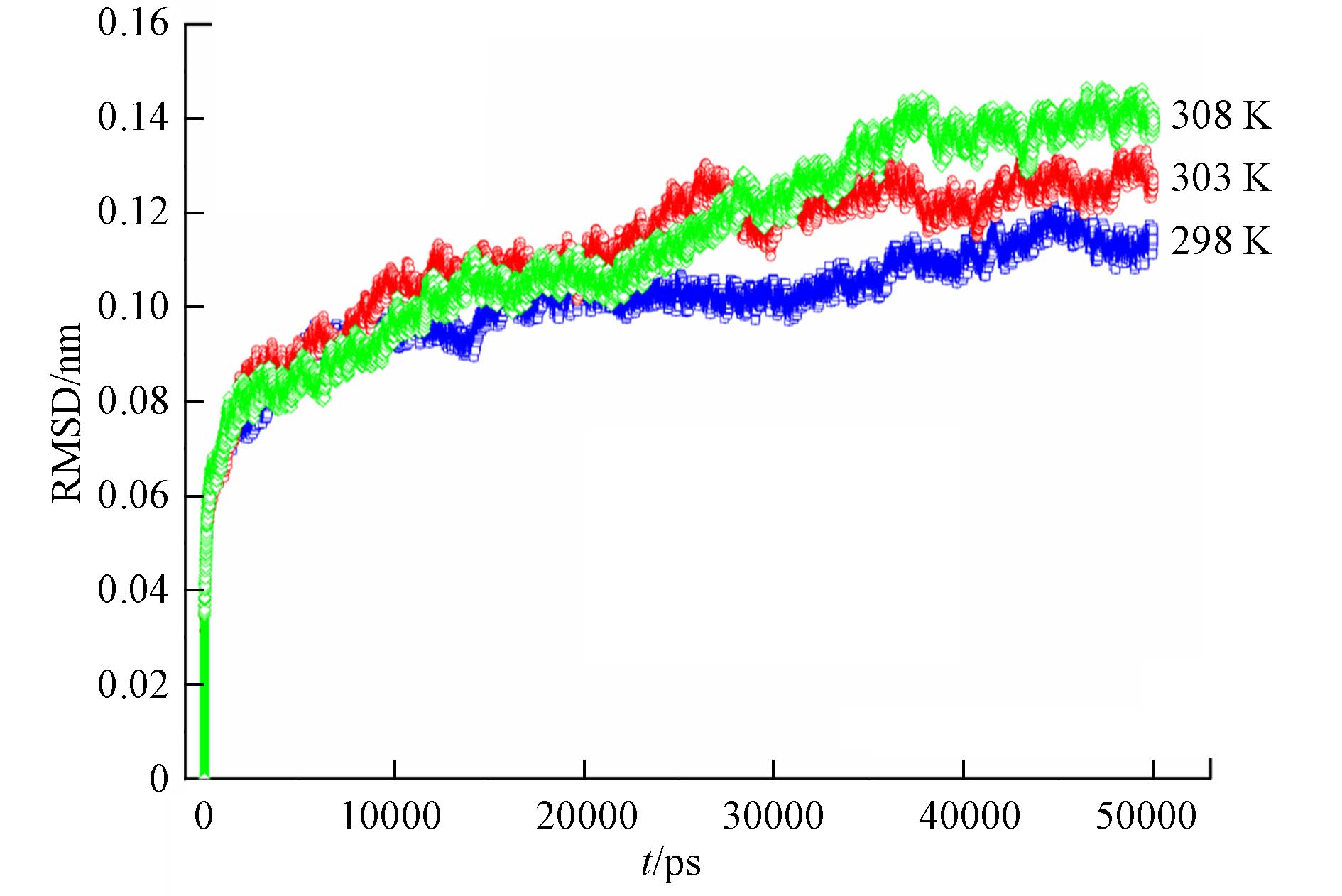
RMSD通常被用来测量酶骨架原子的结构稳定性. RMSD值越高,酶的稳定性就越差. 为了观察Lac与HPAM-3的结合稳定性,计算RMSD值随模拟时间的变化,并以Lac与底物的初始构象作为参考,如图10所示. 当Lac在298 K时与HPAM-3结合时,该酶的骨架振动振幅最小,说明它们在298 K时结合稳定性是最佳的.
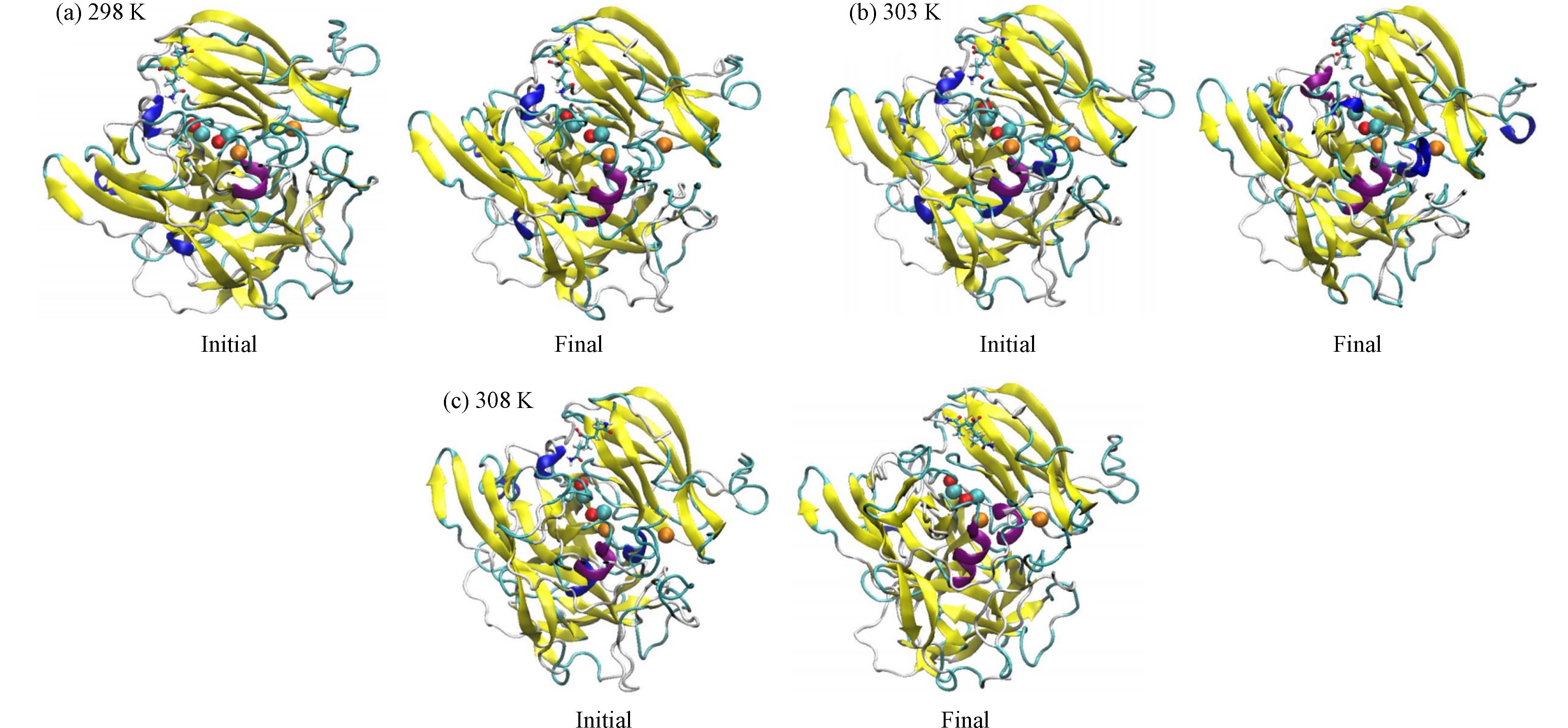
图11显示了MD轨迹采样分析的第一帧和最后一帧. 从图11可以看出,Lac-HPAM-3复合物的构象在298 K和303 K变化不明显,而HPAM-3的取向略有改变,说明该复合物在298 K和303 K下均是稳定的. 但Lac在308 K结合底物时HPAM-3偏离原先的对接位置,推测这可能是由于Lac骨架振动较大导致,即此时的RMSD值最大,因此说明Lac在308 K结合底物时的结合稳定性是最差的.
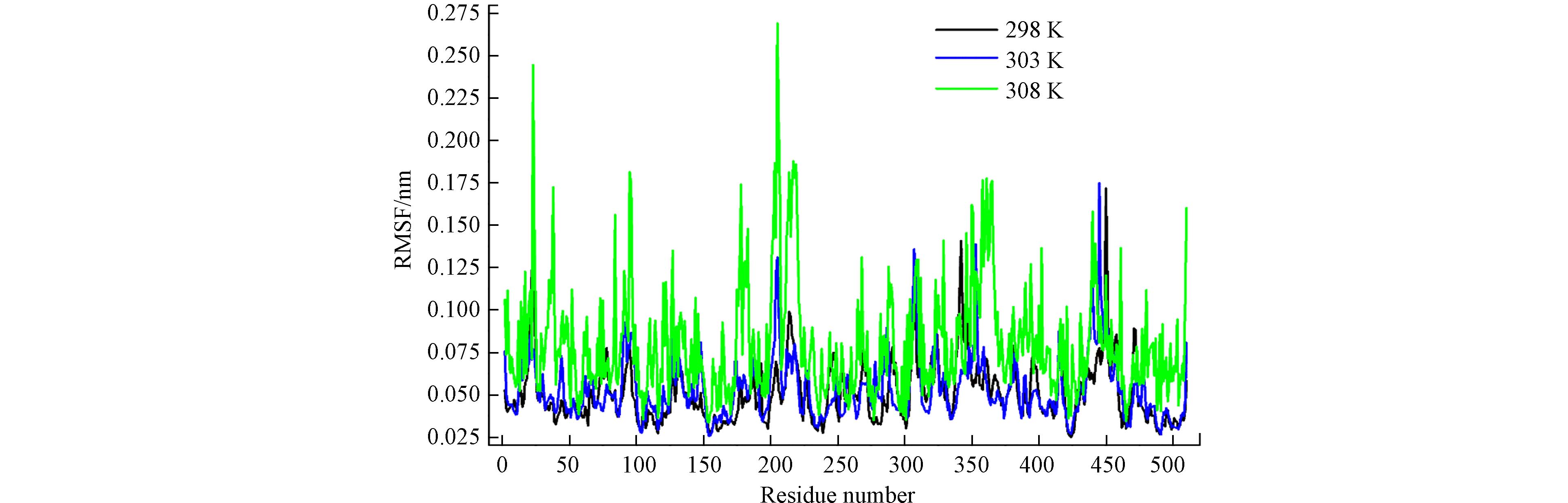
RMSF值越高说明相应的单个残基在酶中的稳定性就越差,而这些残基可能会影响酶结合底物的稳定性. 从图12可以看出,Lac所有残基的RMSF值随温度升高而逐渐增大,而且在298 K下Lac与HPAM-3结合时该酶所有残基的RMSF是最小的,这进一步说明Lac与HPAM-3在298 K时结合是最佳的.
-
采用对接获得了PAA或HPAM与Lac最佳的结合构象,从中选取能量打分皆为正的Lac-底物复合物进行了相互作用分析,然后对最适复合物Lac-HPAM-3分别进行基于亲和力的虚拟突变和不同温度下的MD模拟计算.
(1)通过结合模式分析表明,Lac-HPAM-3享有最高亲和力、最稳定,且它们相互作用最小、结合最好,因此HPAM-3是该酶最适底物;HPAM比PAA更易被Lac降解;Lac对HPAM和PAA的碳链都有一定的耐受,但该酶很难降解分子量较大的聚合物.
(2)通过对打分皆为正的复合物进行相互作用分析可发现,Lac-HPAM-3亲和力最大、最稳定的主要原因是由于形成铵盐; TYR133通过形成氢键以稳定Lac-HPAM复合物,而ARG487一直通过形成疏水以稳定所有复合物.
(3)通过ALA扫描得到,TYR118、TYR133、ARG487和LYS135是Lac降解HPAM-3的关键残基. 通过虚拟饱和突变进一步确定,LYS135和ARG487分别通过形成盐桥和疏水最大程度地影响酶同底物的亲和力.
(4)通过MD模拟计算分析发现,Lac与HPAM-3在298 K时总相互作用能、酶骨架RMSD和酶所有残基RMSF都最小,因此该温度下酶与底物的结合稳定性最佳;在298 K和303 K时复合物构象变化不明显,仅底物取向略有改变,说明酶与底物的结合在298 K和303 K下均是稳定的;308 K时Lac的骨架RMSD最大,导致底物偏离原先对接位置,说明在该温度时酶与底物的结合稳定性是最差的.
枯草芽孢杆菌漆酶与阴离子型聚丙烯酰胺微观作用机理
Microscopic action mechanism of laccase from Bacillus subtilis with anionic polyacrylamide
-
摘要: 阴离子型聚丙烯酰胺(HPAM)常用于煤泥水的澄清处理,产生大量的含聚污水将对选煤厂周边环境造成不利影响. 为探究漆酶降解HPAM的微观作用机理,采用对接模拟了HPAM及其脱氨产物聚丙烯酸(PAA)结构模型与枯草芽孢杆菌漆酶(Lac)的结合,根据-CDOCKER_ Energyscore打分最高原则筛选出Lac的最适底物,然后对该最适复合物分别进行基于亲和力虚拟突变和不同温度下的分子动力学(MD)模拟. 结合模式分析表明,Lac对HPAM-3享有最高亲和力,且其结合最好,因此HPAM-3是该酶的最适底物;HPAM比PAA易被Lac降解;Lac可容纳一定碳链长度的HPAM和PAA. 相互作用分析表明,Lac-HPAM-3亲和力最大主要原因是盐桥;TYR133通过形成氢键以稳定Lac-HPAM-2、Lac-HPAM-3和Lac-HPAM-4,而ARG487通过形成疏水以稳定所有的酶-底物复合物. 基于亲和力虚拟突变分析表明,TYR118、TYR133、ARG487和LYS135是Lac降解HPAM-3的关键残基;LYS135和ARG487分别通过形成盐桥和疏水来最大限度地影响酶同底物的亲和力. MD分析表明,Lac-HPAM-3在298 K时总相互作用能、酶骨架RMSD及所有残基RMSF皆最低,因此该复合物在298 K时结合稳定性最佳;308 K时由于酶骨架RMSD最大,导致底物偏离最初对接位置,因此Lac-HPAM-3在308 K时结合稳定性最差. 这些数据为揭示HPAM酶降解过程奠定基础,为将来突变试验来改造酶提供位点支持.
-
关键词:
- 枯草芽孢杆菌漆酶(Lac) /
- 阴离子型聚丙烯酰胺(HPAM) /
- 对接 /
- 基于亲和力虚拟突变 /
- 分子动力学(MD).
Abstract: Anionic polyacrylamide (HPAM) is often used to clarifying the coal slime water. A large amount of polymer-containing wastewater produced will have adverse effects on the surrounding environment of coal preparation plant. In order to explore the microscopic action mechanism of HPAM by laccase, docking was used to simulate the binding of HPAM and its deamination product polyacrylic acid (PAA) with Bacillus subtilis laccase (Lac). The optimal substrate of Lac was screened according to the highest principle of -CDOCKER_ Energy score. Then, molecular dynamics (MD) simulation at different temperatures and virtual mutation based on affinity were carried out by this optimal complex, respectively. The binding model analysis showed that Lacacquired the highest affinity for HPAM-3, and their binding was the optimal. Hence, HPAM-3 was the most suitable substrate for this enzyme; HPAM was more easily degraded by Lac than PAA; Lac could accommodate HPAM and PAA with a certain carbon chain length. The interaction analysis showed that Lac-HPAM-3 had the highest affinity mainly due to the salt bridge; TYR133 stabilized Lac-HPAM-2, Lac-HPAM-3, and Lac-HPAM-4 by forming hydrogen bonds, while ARG487 stabilized all enzyme-substrate complexes by forming hydrophobicity. The virtual mutation based on affinity analysis showed that TYR118, TYR133, ARG487 and LYS135 in Lac were key residues for degrading HPAM-3; LYS135 and ARG487 affected the affinity of enzyme with substrate to the utmost extent by forming salt bridge and hydrophobicity, respectively. MD analysis showed that the total interaction energy, RMSD of enzyme skeleton and RMSF of all residues were all the lowest for Lac-HPAM-3 at 298 K, so this complex had the optimal binding stability at 298 K; RMSD of the enzyme skeleton was the maximum at 308 K causing the substrate deviated from the initial docking position. Hence, Lac-HPAM-3 had the worst binding stability at 308 K. These data laid the foundation for revealing the enzymatic degradation process of HPAM, and provided site support for mutagenesis tests to modify the enzyme in the future. -

-
表 1 通过CDOCKER工具获得的Lac与底物对接结果
Table 1. 1Docking results of Lac with substrate obtained from CDOCKER protocol
酶-底物复合物
Enzyme-substrate complex负总能打分/ (kcal·mol−1)
-CDOCKER_Energy score负相互作用能打分/ (kcal·mol−1)
-CDOCKER_ Interaction_Energy scoreLac-HPAM-2 24.2360 24.9384 Lac-HPAM-3 31.8442 35.4626 Lac-HPAM-4 28.8940 33.1487 Lac-HPAM-5 −109.688 7.02389 Lac-PAA-2 24.8628 30.3709 Lac-PAA-3 18.8658 33.4581 Lac-PAA-4 17.5539 34.5526 Lac-PAA-5 −152.526 − 25.3122 表 2 Lac与底物的相互作用残基
Table 2. Interaction residues of Lac with substrate
酶-底物复合物a
Enzyme-substrate complex a氢键(数量):总数
H-bond
(number): total疏水(数量):总数
Hydrophobic
(number): total静电吸引(数量):总数
Electrostatic attract
(number): total盐桥(数量):总数
Salt bridge
(number): totalLac-HPAM-2 TYR133(1), LYS135(2): 3 ARG487(2), TYR118(1), TYR133(1): 4 LYS135(1): 1 Lac-HPAM-3 TYR133(1), ASP507(1): 2 ARG487(2): 2 LYS135(1): 1 Lac-HPAM-4 TYR133(1), LYS135(1): 2 ARG487(1): 1 LYS135(1): 1 Lac-PAA-2 ARG487(2), TYR118(1), TYR133(1): 4 LYS135(2): 2 Lac-PAA-3 LYS135(1): 1 ARG487(1), TYR118(1): 2 LYS135(2): 2 Lac-PAA-4 LYS135(1), GLY486(1): 2 ARG487(1): 1 LYS135(2): 2 注:指能量得分皆为正的最佳对接复合物.
Refers to the best docking complex with positive energy score.表 3 Lac-HPAM-3的ALA扫描的结果
Table 3. Results of ALA scan in Lac-HPAM-3
索引
Index突变
Mutation突变能/ (kcal·mol−1)
Mutation energy突变的影响
Effect of mutation1 ASP114ALA −0.04 中性 2 ASP113ALA −0.19 中性 3 ASP116ALA −0.12 中性 4 GLY486ALA −0.01 中性 5 ILE508ALA 0.02 中性 10 ASP507ALA 0.40 中性 11 TYR118ALA 0.57 不稳定 12 TYR133ALA 0.90 不稳定 13 ARG487ALA 1.35 不稳定 14 LYS135ALA 1.62 不稳定 注:表中仅报告了5个最低能量及5个最高能量的突变.
Table reports up to 5 lowest energy and up to 5 highest energy mutations.表 4 Lac-HPAM-3的虚拟饱和突变结果
Table 4. Results of virtual saturation mutation in Lac-HPAM-3
索引
Index突变
Mutation突变能/ (kcal·mol−1)
Mutation energy突变的影响
Effect of mutation1 TYR118ARG −0.36 中性 2 TYR133LYS −0.35 中性 3 TYR133ARG −0.22 中性 4 LYS135LYS −0.18 中性 5 TYR118TRP −0.18 中性 72 TYR118LYS 2.42 不稳定 73 LYS135ASP 2.50 不稳定 74 LYS135TYR 2.52 不稳定 75 LYS135HIS 3.13 不稳定 76 ARG487PRO 4.64 不稳定 注:表中仅报告了5个最低能量及5个最高能量的突变.
Table reports up to 5 lowest energy and up to 5 highest energy mutations. -
[1] 袁亮, 张平松. 煤炭精准开采地质保障技术的发展现状及展望[J]. 煤炭学报, 2019, 44(8): 2277-2284. YUAN L, ZHANG P S. Development status and prospect of geological guarantee technology for precise coal mining[J]. Journal of China Coal Society, 2019, 44(8): 2277-2284 (in Chinese).
[2] 张志军, 孟齐, 刘炯天. 选煤水化学: 循环煤泥水系统的水化学性质[J]. 煤炭学报, 2021, 46(2): 614-623. ZHANG Z J, MENG Q, LIU J T. Water chemistry in coal preparation: Water chemistry properties of circulating coal slime water system[J]. Journal of China Coal Society, 2021, 46(2): 614-623 (in Chinese).
[3] 张志军, 庄丽, 刘炯天. 选煤水化学: 水化学性质对颗粒间相互作用的影响[J]. 煤炭学报, 2021, 46(5): 1685-1693. ZHANG Z J, ZHUANG L, LIU J T. Water chemistry in coal preparation: Effect of water chemistry properties on interparticle interaction[J]. Journal of China Coal Society, 2021, 46(5): 1685-1693 (in Chinese).
[4] REN B, MIN F F, CHEN J, et al. Adsorption mechanism insights into CPAM structural units on kaolinite surfaces: A DFT simulation[J]. Applied Clay Science, 2020, 197: 105719. doi: 10.1016/j.clay.2020.105719 [5] MORTIMER D A. Synthetic polyelectrolytes-a review[J]. Polymer International, 1991, 25(1): 29-41. doi: 10.1002/pi.4990250107 [6] HANSDAH P, KUMAR S, MANDRE N R. Optimization of settling characteristics of coal fine tailings with an anionic polyacrylamide using response surface methodology[J]. International Journal of Coal Preparation and Utilization, 2021, 41(5): 370-383. doi: 10.1080/19392699.2018.1483354 [7] 闵凡飞, 汪婷, 任豹, 等. APAM在水/高岭石界面吸附行为的试验和分子模拟研究[J]. 中国矿业大学学报, 2022, 51(3): 572-580. doi: 10.3969/j.issn.1000-1964.2022.3.zgkydxxb202203019 MIN F F, WANG T, REN B, et al. Experimental and molecular simulation study of the adsorption behavior of APAM at water/kaolinite interface[J]. Journal of China University of Mining and Technology, 2022, 51(3): 572-580 (in Chinese). doi: 10.3969/j.issn.1000-1964.2022.3.zgkydxxb202203019
[8] GUEZENNEC A G, MICHEL C, BRU K, et al. Transfer and degradation of polyacrylamide-based flocculants in hydrosystems: A review[J]. Environmental Science and Pollution Research, 2015, 22(9): 6390-6406. doi: 10.1007/s11356-014-3556-6 [9] KANG X, XIA Z, CHEN R P, et al. Effects of inorganic cations and organic polymers on the physicochemical properties and microfabrics of kaolinite suspensions[J]. Applied Clay Science, 2019, 176: 38-48. doi: 10.1016/j.clay.2019.04.024 [10] SEYBOLD C D. Polyacrylamide review: Soil conditioning and environmental fate[J]. Communications in Soil Science and Plant Analysis, 1994, 25(11/12): 2171-2185. [11] ERKEKOGLU P, BAYDAR T. Acrylamide neurotoxicity[J]. Nutritional Neuroscience, 2014, 17(2): 49-57. doi: 10.1179/1476830513Y.0000000065 [12] JOSHI S J, ABED R M M. Biodegradation of polyacrylamide and its derivatives[J]. Environmental Processes, 2017, 4(2): 463-476. doi: 10.1007/s40710-017-0224-0 [13] 邵振波, 周吉生, 孙刚, 等. 部分水解聚丙烯酰胺驱油过程中机械降解研究: 分子量、粘度及相关参数的变化[J]. 油田化学, 2005, 22(1): 72-77. SHAO Z B, ZHOU J S, SUN G, et al. Studies on mechanical degradation of partially hydrolized polyacrylamide in course of polymer flooding: Changes in relative molecular mass, viscosity and related parameters[J]. Oilfield Chemistry, 2005, 22(1): 72-77 (in Chinese).
[14] ZHOU Y, LI W, WAN W C, et al. W/Mo co-doped BiVO4 for photocatalytic treatment of polymer-containing wastewater in oilfield[J]. Superlattices and Microstructures, 2015, 82: 67-74. doi: 10.1016/j.spmi.2015.02.011 [15] KAY-SHOEMAKE J L, WATWOOD M E, LENTZ R D, et al. Polyacrylamide as an organic nitrogen source for soil microorganisms with potential effects on inorganic soil nitrogen in agricultural soil[J]. Soil Biology and Biochemistry, 1998, 30(8/9): 1045-1052. [16] ZHAO L M, BAO M T, YAN M, et al. Kinetics and thermodynamics of biodegradation of hydrolyzed polyacrylamide under anaerobic and aerobic conditions[J]. Bioresource Technology, 2016, 216: 95-104. doi: 10.1016/j.biortech.2016.05.054 [17] NYYSSÖLÄ A, AHLGREN J. Microbial degradation of polyacrylamide and the deamination product polyacrylate[J]. International Biodeterioration and Biodegradation, 2019, 139: 24-33. doi: 10.1016/j.ibiod.2019.02.005 [18] SANG G L, PI Y R, BAO M T, et al. Biodegradation for hydrolyzed polyacrylamide in the anaerobic baffled reactor combined aeration tank[J]. Ecological Engineering, 2015, 84: 121-127. doi: 10.1016/j.ecoleng.2015.07.028 [19] KAWAI F. Biodegradation of polyethers and polyacrylate[M]//Studies in Polymer Science. Amsterdam: Elsevier, 1994: 24-38. [20] BAO M T, CHEN Q G, LI Y M, et al. Biodegradation of partially hydrolyzed polyacrylamide by bacteria isolated from production water after polymer flooding in an oil field[J]. Journal of Hazardous Materials, 2010, 184(1/2/3): 105-110. [21] SONG T W, LI S S, DING W D, et al. Biodegradation of hydrolyzed polyacrylamide by the combined expanded granular sludge bed reactor-aerobic biofilm reactor biosystem and key microorganisms involved in this bioprocess[J]. Bioresource Technology, 2018, 263: 153-162. doi: 10.1016/j.biortech.2018.04.121 [22] SINGH D, SHARMA K K, JACOB S, et al. Molecular docking of laccase protein from Bacillus safensis DSKK5 isolated from earthworm gut: A novel method to study dye decolorization potential[J]. Water, Air, & Soil Pollution, 2014, 225(11): 2175. [23] LIU Z F, SHAO B B, ZENG G M, et al. Effects of rhamnolipids on the removal of 2, 4, 2, 4-tetrabrominated biphenyl ether (BDE-47) by Phanerochaete chrysosporium analyzed with a combined approach of experiments and molecular docking[J]. Chemosphere, 2018, 210: 922-930. doi: 10.1016/j.chemosphere.2018.07.114 [24] CHEN M, ZENG G M, LAI C, et al. Molecular basis of laccase bound to lignin: Insight from comparative studies on the interaction of Trametes versicolor laccase with various lignin model compounds[J]. RSC Advances, 2015, 5(65): 52307-52313. doi: 10.1039/C5RA07916K [25] ENGUITA F J, MARTINS L O, HENRIQUES A O, et al. Crystal structure of a bacterial endospore coat component: A laccase with enhanced thermostability properties[J]. The Journal of Biological Chemistry, 2003, 278(21): 19416-19425. doi: 10.1074/jbc.M301251200 [26] ZHAO X D, SONG L Z, FU J, et al. Experimental and DFT investigation of surface degradation of polyvinylidene fluoride membrane in alkaline solution[J]. Surface Science, 2011, 605(11/12): 1005-1015. [27] WANG X K, JI G X, HAN X Y, et al. Thiazolidinedione derivatives as novel GPR120 agonists for the treatment of type 2 diabetes[J]. RSC Advances, 2022, 12(10): 5732-5742. doi: 10.1039/D1RA08925K [28] 王方略, 张东晨, 吴学凤, 等. 红球菌酰胺酶降解阴离子型聚丙烯酰胺的亲和力分析[J]. 环境化学, 2023, 42(1): 319-326. doi: 10.7524/j.issn.0254-6108.2021083103 WANG F L, ZHANG D C, WU X F, et al. Affinity analysis of anionic polyacrylamide degraded by amidase from Rhodococcus sp. N-771[J]. Environmental Chemistry, 2023, 42(1): 319-326 (in Chinese). doi: 10.7524/j.issn.0254-6108.2021083103
[29] BERENDSEN H J C, van der SPOEL D, van DRUNEN R. GROMACS: A message-passing parallel molecular dynamics implementation[J]. Computer Physics Communications, 1995, 91(1/2/3): 43-56. [30] DUAN Y, WU C, CHOWDHURY S, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations[J]. Journal of Computational Chemistry, 2003, 24(16): 1999-2012. doi: 10.1002/jcc.10349 [31] BERENDSEN H J C, POSTMA J P M, van GUNSTEREN W F, et al. Interaction models for water in relation to protein hydration[M]//Pullman B. Intermolecular Forces. Dordrecht: Springer, 1981: 331-342. [32] ESSMANN U, PERERA L, BERKOWITZ M L, et al. A smooth particle mesh Ewald method[J]. The Journal of Chemical Physics, 1995, 103(19): 8577-8593. doi: 10.1063/1.470117 [33] TU M L, LIU H X, ZHANG R Y, et al. Analysis and evaluation of the inhibitory mechanism of a novel angiotensin-I-converting enzyme inhibitory peptide derived from casein hydrolysate[J]. Journal of Agricultural and Food Chemistry, 2018, 66(16): 4139-4144. doi: 10.1021/acs.jafc.8b00732 -



 下载:
下载: