-
全程自养脱氮工艺(completely autotrophic nitrogen removal over nitrite,CANON)不但脱氮路径短,污泥产量低,无需外加碳源,比部分硝化-厌氧氨氧化工艺的温室气体排放少[1],还比传统硝化反硝化工艺节省约63%的曝气量,是最具发展前景的高效脱氮工艺[2-3]。然而,其内部主要的脱氮功能菌氨氧化菌(ammonia oxidation bacteria, AOB)和厌氧氨氧化(anaerobic ammonia oxidation, anammox)菌生长缓慢,富集难度大,其活性极易受环境因子(如
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N、$ {\rm{N}}{{\rm{O}}^ -_2} $ -N、DO和pH等)的影响,使得CANON工艺启动和稳定运行面临巨大挑战[4-6]。有研究[7-8]表明,率先接种anammox污泥后,采用厌氧/限氧的运行方式能快速启动CANON工艺。李祥等[9]也指出,采用基于anammox污泥的厌氧/限氧启动模式是一种行之有效的策略,其在升流式生物膜反应器中,比基于普通活性污泥的好氧/限氧运行方式的启动时长缩短约72 d。但有关该启动模式所适用的调控参数,如DO、游离氨(free ammonia, FA)和游离亚硝酸(free nitrous acid, FNA)质量浓度等,依然存在较大争议,亟待进一步探究。DO作为CANON工艺运行的关键环境因子,其最适宜的调控范围至今尚未有定论。有研究者指出,在anammox污泥基础上,接种3 g·L−1的部分硝化污泥,并控制启动前期的DO为0.5~1 mg·L−1,待稳定运行后保持1.5~4.3 mg·L−1的DO范围,可以使CANON工艺高效运行,氮去除率(nitrogen removal rate, NRR)可达1.22 kg·(m3·d)−1[10]。以同样的启动模式,FANG等[11]则认为,调控DO为1.5~2.0 mg·L−1,亦可顺利启动CANON工艺,但NRR仅120 g·(m3·d)−1。另有研究者发现,高于0.7 mg·L−1的DO水平便会使
$ {\rm{N}}{{\rm{O}}^ -_2} $ 过量累积[12],需要严格控制DO为(0.35 ± 0.05) mg·L−1,才可实现AOB和anammox菌的协同生长,使NRR达0.51 kg·(m3·d)−1[13]。此外,$ {\rm{N}}{{\rm{H}}^ +_4} $ -N和$ {\rm{N}}{{\rm{O}}^ -_2} $ -N虽然是AOB和anammox菌的生长基质,但也极易抑制AOB和anammox菌的生长[14-15],主要以FA和FNA形式构成抑制,而且受体系内的pH影响很大[16]。部分研究结果表明,AOB和anammox菌的FNA抑制阈值分别为100 µg·L−1[17-18]和15 µg·L−1[4]。然而,PUYOL等[19]发现,FNA对anammox菌抑制的质量浓度为117 µg·L−1。同时,针对FA的抑制阈值,不同研究结果差异较大。有研究者指出,FA对anammox菌的抑制阈值为20~25 mg·L−1[20];部分研究表明,高于32.5 mg·L−1的FA会严重抑制AOB和anammox菌的生长[17];另有研究者发现,超过90 mg·L−1的FA才会抑制anammox菌活性[21]。以上研究结果充分表明,有关CANON工艺的运行调控策略急需进一步探究。尽管已有研究中均能顺利启动CANON工艺,并应用于实际废水的治理中。但关于CANON工艺启动模式、运行过程中FA和FNA的影响、高负荷运行所需的最佳环境条件及启动运行过程中微生物群落的响应特性仍有必要进行深入研究。为此,本研究以稳定运行的anammox系统为研究对象,先考察适宜anammox菌生长的最佳条件;再通过调控所筛选的环境条件,采取逐渐降低
$ {\rm{N}}{{\rm{O}}^ -_2} $ -N和提升$ {\rm{N}}{{\rm{H}}^ +_4} $ -N质量浓度的方式启动CANON工艺,并探究整个实验过程中微生物群落的响应特性;最后,深入分析CANON启动运行过程中,FA和FNA对氮转化速率的影响,以期为CANON工艺的实际应用提供参考。
全文HTML
-
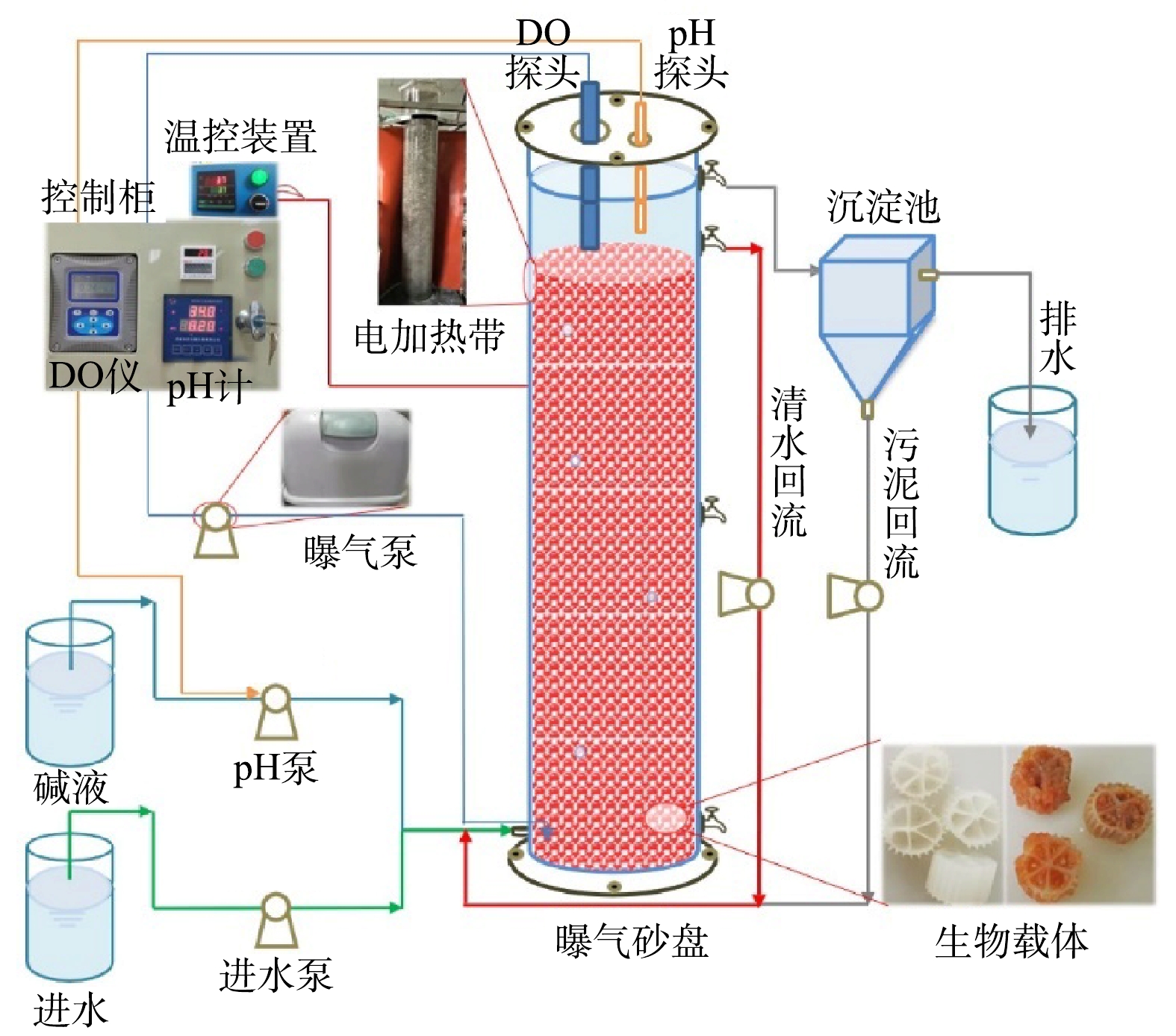
实验采用升流式厌氧固定床反应器,其示意图见图1。反应器有效容积为100 L,内径为30 cm,内部装填鲍尔环填料,顶部加盖,由螺钉和胶条密封,上部安装DO、pH探头并预留排气孔。距底部2 cm和顶部10 cm处分别设有进水口和出水口,中部和底部设有污泥采样口。反应器内的DO和pH分别通过哈希溶氧仪(SC200, HACH Water Quality Analysis Instrument Ltd, USA)和工业在线pH计(pH7203,成都锐新仪器仪表有限公司,中国)在线控制DO为0.2~0.5 mg·L−1和pH 为7.0~7.2[6],水力停留时间为1 d。反应器外部包裹电加热带以确保内部恒温为(35±2) ℃,且避光运行,以维持anammox菌适宜的生长环境。
-
anammox批次实验的污泥取自本实验室已稳定运行180 d,脱氮性能良好的anammox中试系统[15, 22]。该系统在进水
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N和$ {\rm{N}}{{\rm{O}}^ -_2} $ -N质量浓度均为300 mg·L−1时,总氮去除率(nitrogen removal efficiency, NRE)和NRR分别达89.88%和0.64 kg·(m3·d)−1。从中试系统中采集污泥样品,控制温度为35 ℃,通过4个批次实验,分别考察DO、pH、FNA和FA对比厌氧氨氧化活性(specific anammox activity, SAA)的影响,每个处理设置3个生物学重复。其中,不同的DO环境是通过调控敞口和闭口条件下的摇床转速实现;pH是在控制$ {\rm{N}}{{\rm{H}}^ +_4} $ -N和$ {\rm{N}}{{\rm{O}}^ -_2} $ -N的质量浓度不变的条件下,利用HCl和NaOH调节;FNA和FA是在控制pH不变的条件下,分别调整$ {\rm{N}}{{\rm{O}}^ -_2} $ -N和$ {\rm{N}}{{\rm{H}}^ +_4} $ -N的质量浓度得以实现;pH、FNA和FA对SAA的影响实验均是在闭口(厌氧)条件下进行,具体运行参数如表1所示。另外,SAA测定的具体操作过程参考先前的研究中的方法[15, 22]。 -
在中试anammox系统内,接种20 L混合液悬浮固体(mixed liquor suspended solids, MLSS)和混合液挥发性悬浮固体(mixed liquor volatile suspended solids, MLVSS)分别为7.05 g·L−1和4.19 g·L−1的城市污水处理厂活性污泥,开启曝气,按照表2的操作条件启动CANON工艺。启动过程共经历3个阶段:适应阶段Ⅰ(0~69 d)、活性提高阶段Ⅱ(70~149 d)和稳定运行阶段Ⅲ(150~162 d)。运行期间,采集进出水样品经0.45 μm滤膜过滤后,分别采用纳氏试剂法、N-(1-萘基)乙二胺分光光度法和紫外分光光度法测定样品中的
$ {\rm{N}}{{\rm{H}}^ + _4}$ -N、$ {\rm{N}}{{\rm{O}}^ -_2} $ -N和$ {\rm{N}}{{\rm{O}}^ -_3} $ -N[23];污泥样品MLSS、MLVSS均按标准方法[24]测定。 -
1) DNA提取。在CANON工艺启动过程中,采集0、18、44、69、77、83、89、98、131、139、143、149和162 d的污泥样品,经30 min静沉后,称取500 mg污泥样品,使用FastDNATM SPIN Kit for Soil (LLC, MP Biomedicals, USA)提取试剂盒,进行DNA提取。DNA经1%琼脂糖凝胶电泳和Nanodrop (ND1 000, Gene Company Limited, China)进行质检后,−80 ℃保存,用于后续分析。
2)实时定量PCR。选取0、18、44、69、77、83、89、98、131、139、143和149 d的DNA样品,采用Roche LightCycler® 480 Ⅱ (Roche Diagnostics Ltd, Rotkreuz, Swltzerland)实时荧光定量系统进行qPCR分析,以定量测定反应体系内anammox菌、AOB、反硝化菌(denitrifying bacteria, DNB)和亚硝酸盐氧化菌(nitrite oxidation bacteria, NOB)中氮转化功能基因拷贝数。qPCR的反应体系及操作过程参考先前的研究[22],采用的特异性引物见表3。
3) Illumina高通量测序。选用引物515F (5′-GTG CCA GCM GCC GCG G-3′)和907R (5′-CCG TCA ATT CMT TTR AGT TT-3′)对0、77、139和162 d的DNA样品进行扩增,扩增产物经2%琼脂糖凝胶电泳质控,经均一化后,进行Miseq文库构建,并采用Illumina Miseq测序平台对样品进行高通量测序。
-
根据式(1)和式(2)[32]计算CANON工艺运行中FA和FNA的质量浓度。本研究中,整个实验过程没有添加有机碳源,故反应器内的脱氮过程主要以anammox途径为主导。系统中亚硝化和anammox途径对氨氮的转化率由式(3)~式(5)[33-35]计算。由以上的化学计量关系计算以anammox途径为主导的NRR、氨氧化速率(ammonium oxidation rate,AOR)和NOB的硝化速率(nitrite oxidation rate,NOR),如式(6)~式(8)[33-35]所示。
式中:
$ {C}_{{\rm{F}}{\rm{A}}} $ 、$ {C}_{{\rm{F}}{\rm{N}}{\rm{A}}} $ 、$ {C}_{{{\rm{N}}{\rm{H}}}_{4}^{+}{\text{-N}}} $ 和$ {C}_{{{\rm{N}}{\rm{O}}}_{2}^{-}{\text{-N}}} $ 分别为FA、FNA、$ {\rm{N}}{{\rm{H}}^ +_4} $ -N和$ {\rm{N}}{{\rm{O}}^ - _2}$ -N的质量浓度,mg·L−1;T为温度,K。式中:
$ {V}_{{\rm{N}}{\rm{R}}{\rm{R}}} $ 、$ {V}_{{\rm{A}}{\rm{O}}{\rm{R}}} $ 、$ {V}_{{\rm{N}}{\rm{O}}{\rm{R}}} $ 分别为NRR、AOR和NOR,kg·(m3·d)−1;$ {\Delta C}_{{{\rm{N}}{\rm{H}}}_{4}^{+}{\text{-N}}} $ 、$ {\Delta C}_{{{\rm{N}}{\rm{O}}}_{3}^{-}{\text{-N}}} $ 和$ {\Delta C}_{{\rm{T}}{\rm{N}}} $ 分别为进出水中$ {\rm{N}}{{\rm{H}}^ + _4}$ -N、$ {\rm{N}}{{\rm{O}}^ -_3} $ -N和TN的质量浓度差,mg·L−1。
1.1. 实验装置和运行条件
1.2. anammox批次实验
1.3. CANON工艺启动
1.4. DNA提取、实时定量PCR与多样性测定
1.5. 数据处理
-
SAA是anammox菌转化
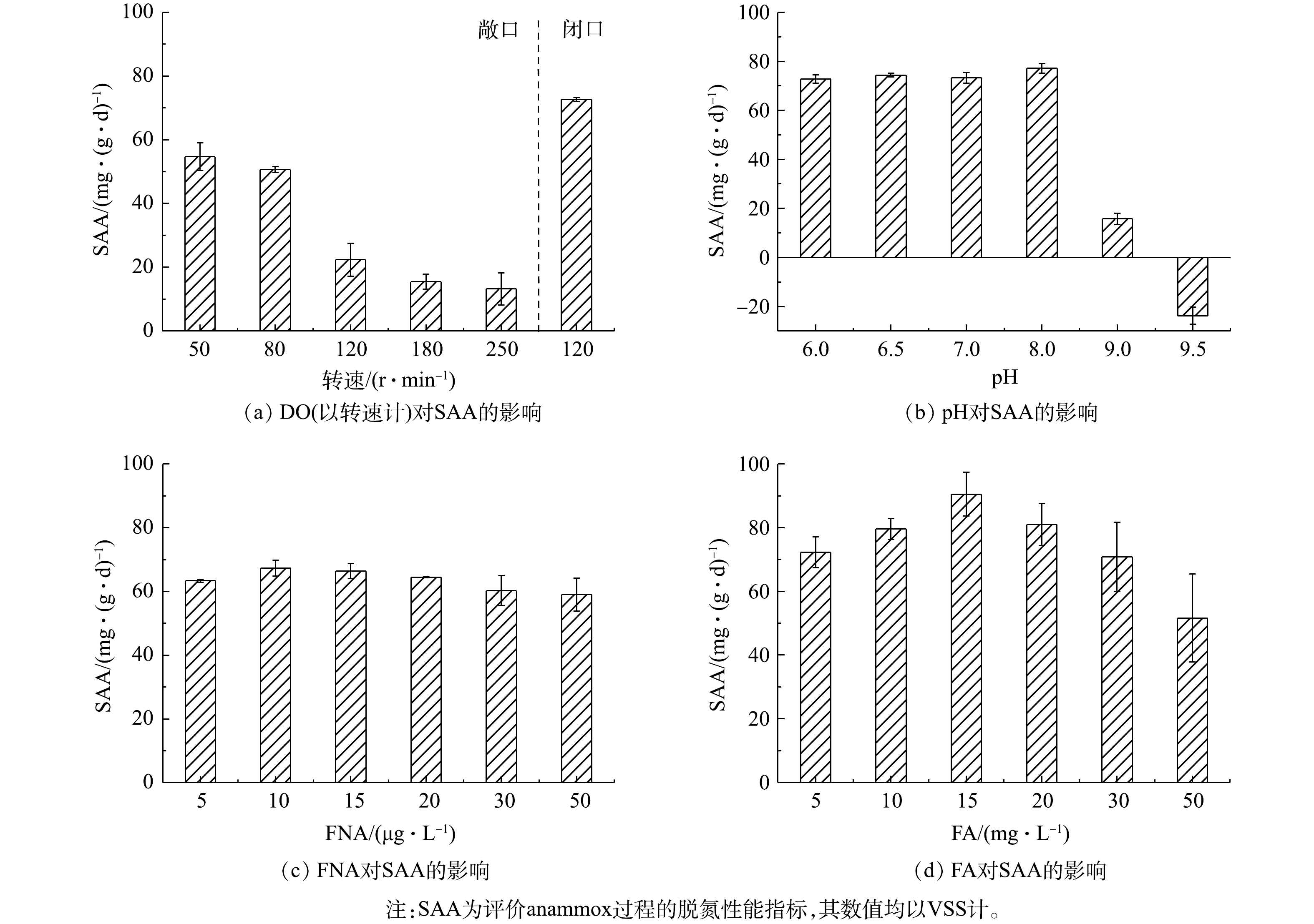
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N和$ {\rm{N}}{{\rm{O}}^ -_2} $ -N为N2的反应速率,常被作为评价anammox过程的脱氮性能的指标,其数值均以VSS计[36]。为探究由anammox转为CANON工艺的最佳转化条件,首先通过批次实验考察了不同DO、pH、FNA和FA对SAA的影响。由图2可知,SAA随敞口环境下转速的增加而逐渐降低,与DO环境有显著的负相关关系(P<0.01),且当转速达120 r·min−1 (DO约0.78 mg·L−1)时,SAA迅速下降至22.39 mg·(g·d)−1(均以VSS计),较80 r·min−1时(DO为0.4~0.5 mg·L−1)下降55.79%。这说明CANON工艺启动过程中应控制DO≤0.5 mg·L−1,以确保anammox菌具有较高的活性。同时,当pH为6.0~8.0时,SAA随pH增加而逐渐升高,当pH为8.0时,SAA达到最高值77.16 mg·(g·d)−1;之后,SAA急剧下降,当pH为9.0时,SAA仅为15.71 mg·(g·d)−1,较pH为8.0时下降了79.64%。FNA和FA对SAA均有低浓度促进、高浓度抑制的作用,且FNA和FA分别在10 µg·L−1和15 mg·L−1时,SAA达到峰值,分别为67.33 mg·(g·d)−1和90.52 mg·(g·d)−1。当高于该阈值后,FNA对SAA抑制效果并不明显,而FA与SAA则呈显著的负相关关系(P<0.01)。该结果与LI等[17]和YANG等[15]的研究结果一致。LI等[17]指出32.5 mg·L−1的FA会显著抑制anammox菌活性。YANG等[15]也指出控制FNA<15 μg·L−1、FA<15 mg·L−1,可以快速恢复经强碱冲击后严重恶化的anammox过程。以上结果表明,在DO≤0.5 mg·L−1、pH为7.0~8.0、FNA 为5~20 µg·L−1和FA为10~20 mg·L−1的条件下,有利于高活性anammox菌的生长。 -
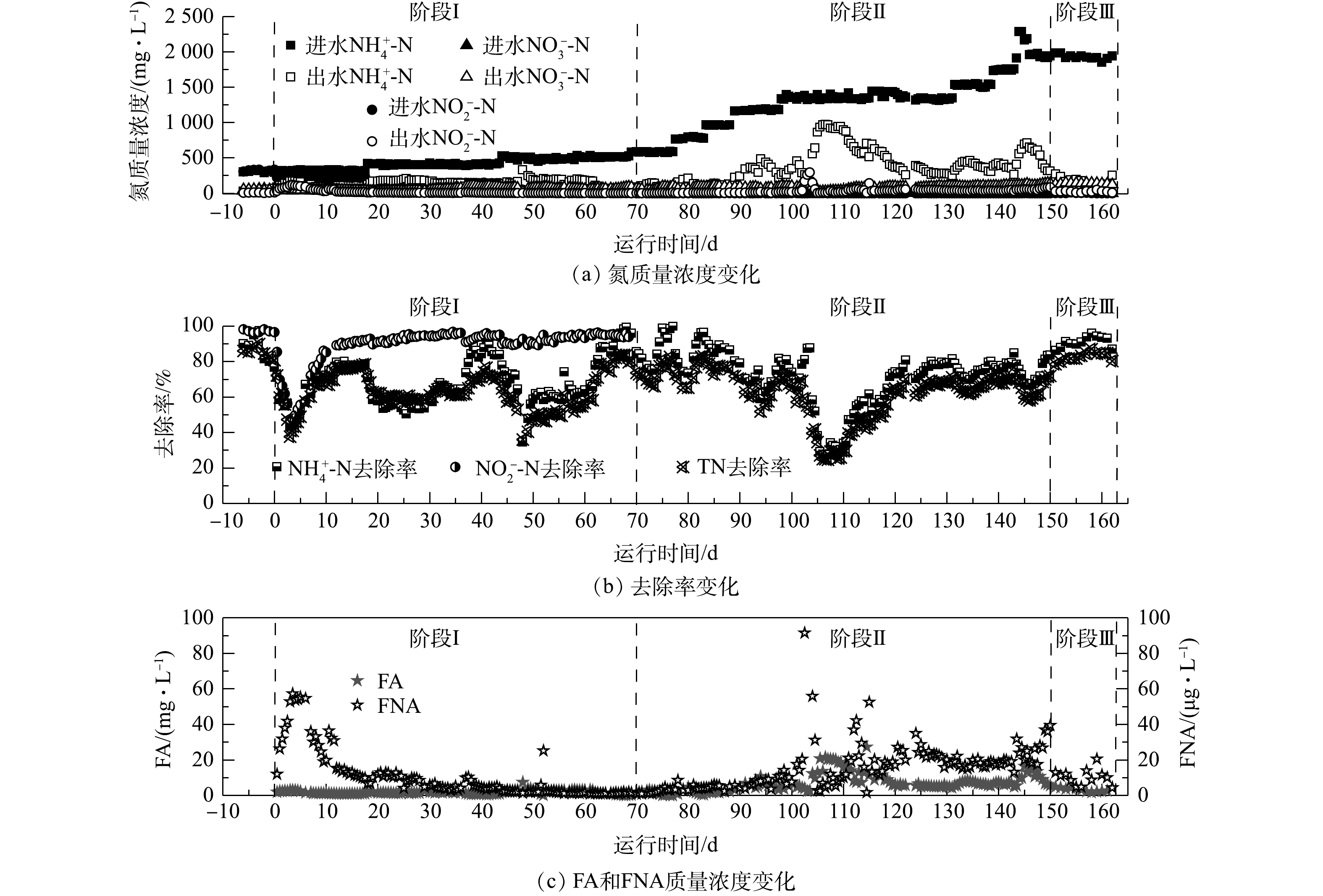
图3反映了CANON工艺启动过程中的水质变化。由图3可知,CANON工艺的启动经历了3个阶段:适应阶段(阶段Ⅰ,0~69 d)、活性提高阶段(阶段Ⅱ,70~149 d)和稳定运行阶段(阶段Ⅲ,150~162 d)。在阶段Ⅰ中,在开始曝气后的1~3 d,DO约0.40 mg·L−1,反应器脱氮性能急速恶化,NRE由开始曝气前的80.78%迅速降至37.73%,出水
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N和$ {\rm{N}}{{\rm{O}}^ - _2}$ -N质量浓度也较开始曝气前分别增加了3.16倍和21.14倍;但随后DO逐步稳定于0.27 mg·L−1,反应器内微生物也逐渐适应曝气环境,第17天时,NRE已恢复至78.56%,在第18天和44天,在2次提高进水基质中质量浓度后,也观察到该现象。这是因为,在提高进水负荷的同时增加曝气量,DO质量浓度也随之提高(0.51 mg·L−1),可抑制anammox菌活性。之后由于AOB等好氧菌的快速增殖:一方面消耗内部DO,使其降低至0.3 mg·L−1以下;另一方面促进生物膜或颗粒污泥的形成,进而削弱DO的抑制,从而使anammox体系表现一定的适应性[37]。有研究[38-39]表明,在CANON工艺启动的适应阶段,反应器内无$ {\rm{N}}{{\rm{O}}^ -_3} $ 的明显累积,且0.2~0.5 mg·L−1的微氧环境有利于AOB的增殖和NOB的抑制,同时仍会存在一定程度的反硝化作用。在阶段Ⅱ中,停止
$ {\rm{N}}{{\rm{O}}^ -_2} $ 的添加,并通过逐渐增加$ {\rm{N}}{{\rm{H}}^ +_4} $ -N的进水质量浓度以提升CANON工艺的进水氮负荷,发现该阶段anammox菌生长所需的$ {\rm{N}}{{\rm{O}}^ - _2}$ -N完全可以由AOB提供,且NRE一直维持在70.12%左右,无$ {\rm{N}}{{\rm{O}}^ -_2} $ -N和$ {\rm{N}}{{\rm{O}}^ -_3} $ -N的明显累积现象。仅在第102 d时,由于DO在线控制器故障,持续过量曝气约12 h,导致污泥大量流失,DO达6.84 mg·L−1左右。此时,FNA质量浓度率先剧增至150.56 mg·L−1,随后于1 d内降至10 µg·L−1以下,而FA质量浓度却渐增至26.52 mg·L−1,并持续约5 d,导致NRE由70.10%迅速降至24.69%,出水$ {\rm{N}}{{\rm{H}}^ +_4} $ -N和$ {\rm{N}}{{\rm{O}}^ -_2} $ -N分别达到965.6 mg·L−1和14.3 mg·L−1。但经过约14 d的调整,控制DO约为0.22 mg·L−1,FA为5~20 mg·L−1,反应器在122 d时NRE已恢复至70.14%。上述结果表明,由anammox向CANON工艺转化的过程受DO和FA影响较大,需要控制DO为0.2~0.5 mg·L−1,FA为5~20 mg·L−1。YANG等[15]指出,CANON工艺在强碱冲击条件下,通过控制FA<15 mg·L−1,可以快速恢复其中的anammox过程。在阶段Ⅲ中,在进水$ {\rm{N}}{{\rm{H}}^ +_4} $ -N约为2 000 mg·L−1的情况下,反应器$ {\rm{N}}{{\rm{H}}^ +_4} $ -N去除率和NRE分别稳定在91.21%和82.77%左右。该结果远优于有关文献报道的结果[40-41],这说明CANON工艺已顺利启动运行。 -
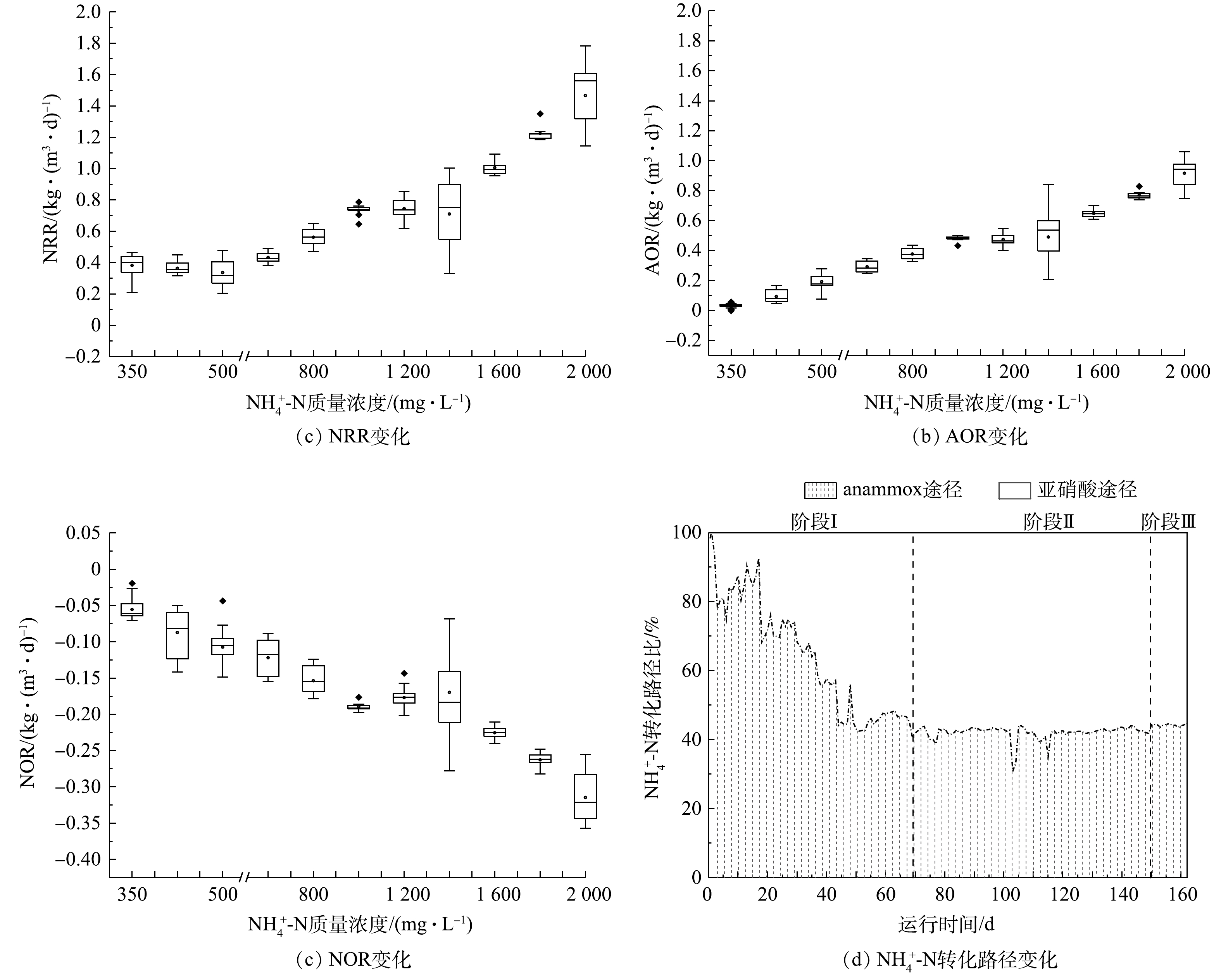
1) CANON工艺启动过程中氮转化路径变化。由图4可看出,在阶段Ⅰ,随着
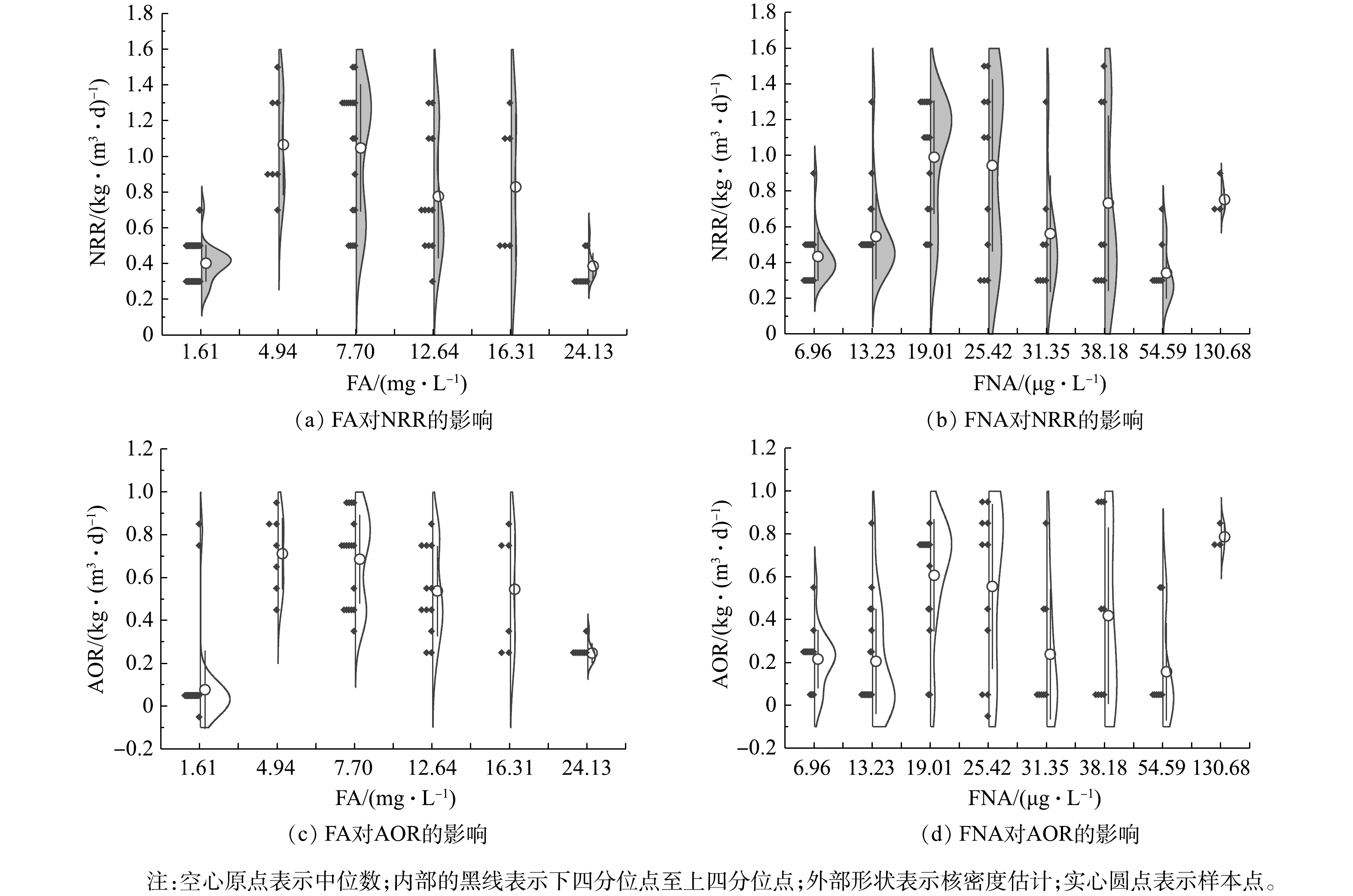
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N质量浓度的提升,NRR始终稳定于(0.47 ± 0.04) kg·(m3·d)−1,而AOR则逐渐增加至0.28 kg·(m3·d)−1。同时,由图4(d)可看出,该阶段通过亚硝化过程转化$ {\rm{N}}{{\rm{H}}^ +_4} $ -N的路径比自开始曝气逐渐增加,至适应阶段末,anammox和亚硝化的$ {\rm{N}}{{\rm{H}}^ +_4} $ -N转化路径比稳定于0.85左右。这是适应阶段CANON工艺中微生物演替的结果。在阶段Ⅱ,NRR和AOR均随$ {\rm{N}}{{\rm{H}}^ +_4} $ -N质量浓度的提升而逐渐增加,相应地分别从0.47 kg·(m3·d)−1和0.29 kg·(m3·d)−1逐渐增加到1.37 kg·(m3·d)−1和0.94 kg·(m3·d)−1,于稳定阶段NRR达1.65 kg·(m3·d)−1。但由图(4)可以看出,$ {\rm{N}}{{\rm{H}}^ +_4} $ -N质量浓度为1 400 mg·L−1时,NRR和AOR数据较分散,正是因为该阶段曝气装置故障所致。在阶段Ⅱ,anammox和亚硝化途径对$ {\rm{N}}{{\rm{H}}^ +_4} $ -N的转化路径比依然稳定在0.73左右。另外,整个实验过程中,NOR都为负值,且呈现逐渐下降的状态,表明0.2~0.5 mg·L−1的DO条件,可以很好地实现对NOB的抑制,但仍有部分反硝化作用存在[38]。2) CANON工艺启动过程中FA和FNA对氮转化速率的影响。为进一步探究FA和FNA对CANON工艺中氮转化速率的影响,本研究选取CANON启动过程中适应阶段和故障阶段的运行数据,按照FA (0~2、3~5、6~10、11~15、15~20、> 20 mg·L−1)和FNA (1~10、11~15、16~20、21~30、31~35、35~40、41~60、> 60 µg·L−1)的实际质量浓度进行区间划分,经深入分析后绘制小提琴图(见图5)。由图5(a)和图5(c)可看出,FA对NRR和AOR均起到低浓度促进、高浓度抑制的作用。在FA<10 mg·L−1时,NRR和AOR均随FA的增加而增加,且呈现显著的相关性(P<0.01);而当FA>10 mg·L−1时,则NRR和AOR均与FA呈现负相关关系(P<0.01)。但NRR和AOR在FA为15~20 mg·L−1的环境中,分别较FA为6~10 mg·L−1时平均下降了20.92%和20.55%;而在FA>20 mg·L−1的环境中,NRR和AOR却较FA为15~20 mg·L−1时的相应值分别下降了53.38%和54.68%。该结果与FA对anammox菌活性影响的批次结果相近,表明在实际运行过程中,FA为5~20 mg·L−1的环境有利于anammox向CANON工艺的转化。此外,如图5(b)和图5(d)所示,当FNA≤20 µg·L−1时,NRR和AOR均随FNA质量浓度增加而增加(P<0.01);而当FNA>20 µg·L−1时,则与NRR和AOR无显著的相关关系(P>0.05)。anammox批次实验、anammox向CANON工艺转化过程中水质变化以及氮转化速率分析结果表明,将FA调控为10~20 mg·L−1,可促进anammox向CANON工艺的转化。
-
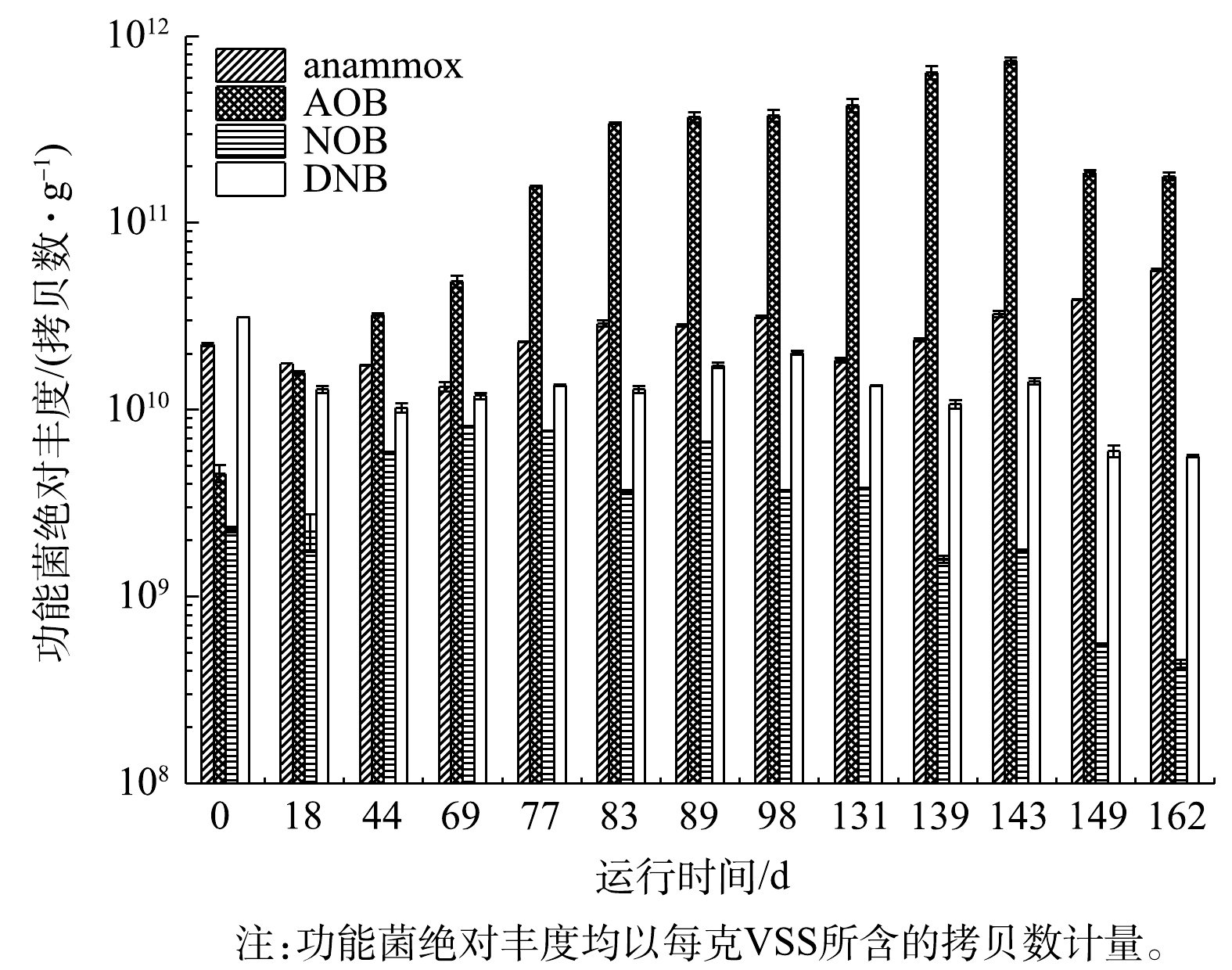
由图6可见,AOB丰度随CANON工艺的启动呈先上升后下降的趋势,并于143 d时达到峰值,即7.83×1011拷贝数·g−1 (均以VSS计),之后因
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N质量浓度升高至1 800 mg·L−1以上,导致AOB丰度有所下降,表明高质量浓度的$ {\rm{N}}{{\rm{H}}^ +_4} $ -N对AOB具有一定的抑制作用。LI等[17]的研究结果表明,高于32.5 mg·L−1的FA会严重抑制AOB的生长。也有研究结果[2, 4, 15]表明,相较于AOB,高质量浓度的$ {\rm{N}}{{\rm{H}}^ +_4} $ -N更适合于anammox菌的生长,使2种菌的氮转化过程达到平衡状态,因此,CANON工艺多应用于高浓度$ {\rm{N}}{{\rm{H}}^ + _4}$ -N废水的治理。而anammox菌丰度在CANON工艺启动过程中出现2次下降。首先在0~69 d,随着开始曝气,anammox菌活性受到抑制,丰度逐渐下降至1.33×1010拷贝数·g−1;其次,在98~131 d,因曝气装置故障,污泥大量流失,导致anammox菌丰度再次下降了41.31%,但随后丰度逐渐增加并稳定在4.41×1010拷贝数·g−1。尽管anammox菌丰度与AOB丰度相差1个数量级,但CANON工艺的NRR一直处于提升状态,且anammox和亚硝化路径对$ {\rm{N}}{{\rm{H}}^ + _4}$ -N的转化比稳定在0.73左右。这可能是因为AOB分泌的代谢物促进了anammox菌代谢酶的活性,使其在低丰度条件下呈现高的脱氮活性[42-44]。至于NOB,其丰度仅在适应阶段(0~69 d) 因开启曝气和较低质量浓度的FA抑制而有所增加,但随后便逐渐降低至4.38×108拷贝数·g−1左右。这可能是反应器长期保持低DO,且随着$ {\rm{N}}{{\rm{H}}^ +_4} $ -N质量浓度增加,FA对NOB的抑制增强所致[17-18]。统计分析结果表明,NOB丰度与FA质量浓度呈显著的负相关关系(P<0.05),而与FNA无显著相关性(P>0.05)。这与LI等[17]的研究结果一致,他们指出10~17 mg·L−1的FA便会对NOB产生明显的抑制作用。而DNB则如预期一样,随着CANON工艺启动,丰度逐渐下降,最终仅为5.64×109拷贝数·g−1[6]。 -
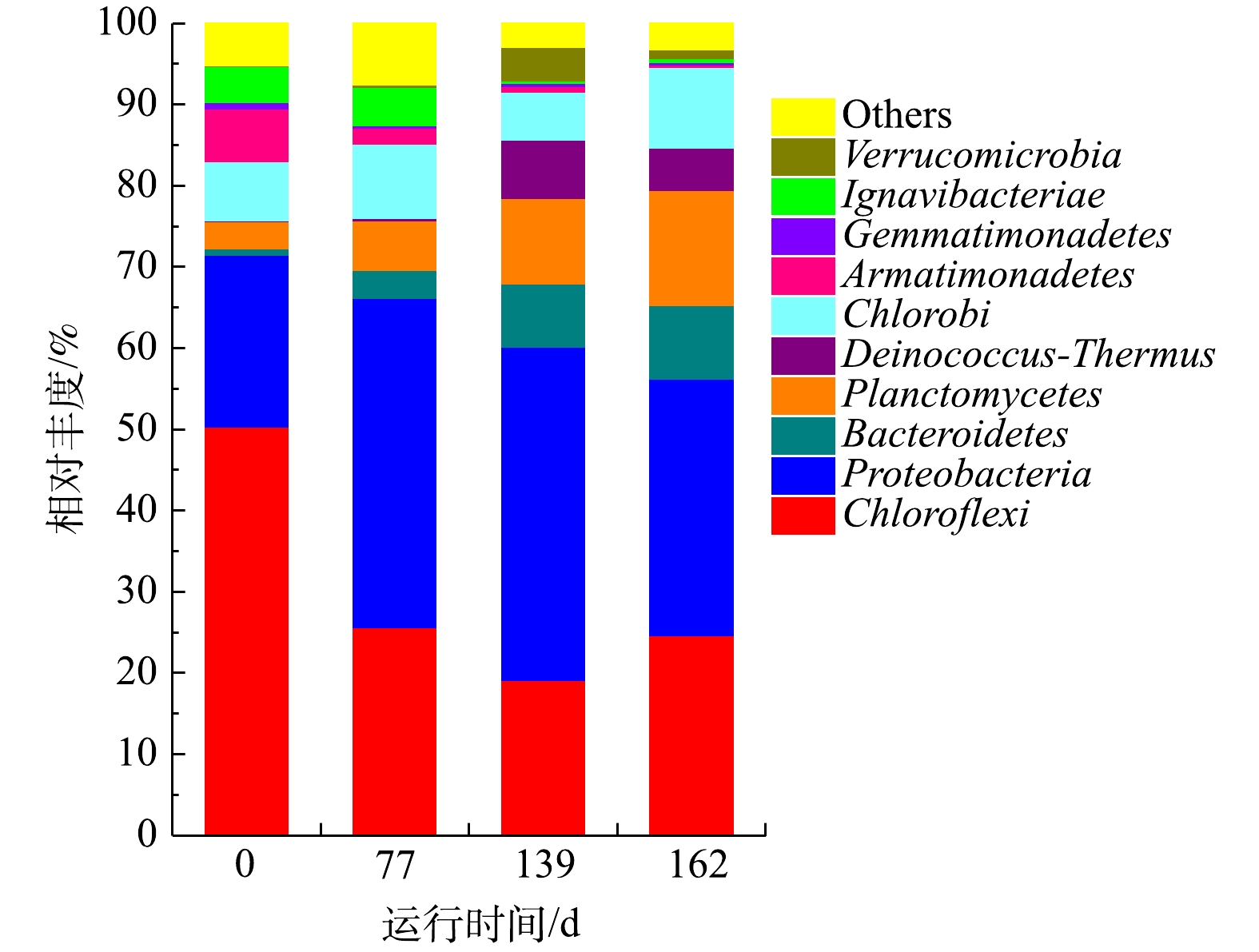
1)门水平物种的丰度分析。图7显示,CANON工艺启动过程以Chloroflexi、Proteobacteria、Bacteroidetes、Planctomycetes等为优势菌门。其中,Chloroflexi可以降解碳水化合物和细胞代谢物,在脱氮系统中对稳定颗粒结构起关键作用[45-46]。Proteobacteria是参与脱氮的主要菌群之一,包括AOB、NOB和DNB [46-47]。Bacteroidete是废水处理、厌氧消化污泥和土壤中普遍存在的菌门[48-49],可以转化细胞代谢所产生的有机物为CO2,为anammox菌提供无机碳源[50-51]。Chloroflexi、Proteobacteria和Bacteroidetes均是常见的与anammox菌共存的菌群[52]。而Planctomycetes则是anammox菌所在的菌门[47]。从测序结果来看,在CANON工艺启动过程中,Planctomycetes和Bacteroidetes丰度呈现逐渐上升的趋势,最终分别达9.04%和14.16%,这是NRR不断获得提升的重要原因。Chloroflexi丰度则呈现先下降后上升的趋势,于139 d时导到最低,而Proteobacteria丰度与之完全相反。这可能是因为曝气后,系统DO升高引起部分厌氧微生物失活,从而释放胞内物或分泌大量EPS等物质来保护自己[53-54],Chloroflexi刚好以此为碳源生长。之后,随着工艺的运行,这些物质被逐渐淘洗掉,而此时Proteobacteria中的AOB和NOB等好氧微生物适宜在该DO环境中生长,故而导致Chloroflexi丰度下降、Proteobacteria丰度增加的趋势。直到后期由于
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N质量浓度过高,再次引起微生物的淘洗和AOB的抑制,使两菌门丰度变化趋势互换。这与qPCR的结果相一致。CHEN等[55]发现,Chloroflexi会优先利用衰变细菌的代谢物,并有助于COD的去除。2)属水平物种的丰度分析。表4展示了CANON启动过程中前5种优势菌属和部分AOB、DNB和anammox菌属的相对丰度。CANON工艺启动过程中,以Nitrosomonas、SM1A02、Unidentified_Anaerolineaceae、Truepera和Arenimonas为优势菌属。Nitrosomonas作为AOB中的优势菌属,可以消耗
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N和DO,为DNB和anammox菌创造厌氧环境[44, 50]。有研究结果表明,SM1A02具有anammox能力,可能是一种新的anammox菌株,与Candidatus Jettenia、Brocadia和Kuenenia共同构成anammox菌群[42, 44, 56]。属层面的物种多样性结果显示,CANON启动过程中,AOB相对丰度先增加后降低,于第139天达到峰值23.42%。Anammox菌相对丰度则一直处于上升的趋势,该结果与相同时期的qPCR结果相一致,进一步解释了水质结果中NRR一直处于上升趋势的原因。属层面DNB相对丰度结果也与qPCR检测结果相同,一直处于下降趋势。所有这些检测结果均表明CANON工艺已成功启动。
2.1. 环境条件对anammox菌活性的影响
2.2. CANON工艺启动过程中水质变化
2.3. CANON工艺启动过程中氮转化路径变化及FA和FNA对氮转化速率的影响
2.4. CANON工艺启动中氮转化功能菌丰度变化
2.5. CANON工艺启动中微生物群落变化
-
1) DO≤0.5 mg·L−1、pH为7.0~8.0、FNA为5~20 µg·L−1和FA为10~20 mg·L−1的环境条件有利于anammox菌的活性维持;控制DO为0.2~0.5 mg·L−1、pH为7.0~7.2和FA为10~20 mg·L−1,可以有效抑制NOB生长,促使anammox工艺顺利转为CANON工艺,使其在进水
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N为2 000 mg·L−1的条件下,实现平均82.77%的NRE和最高1.67 kg·(m3·d)−1的NRR。2) 在CANON工艺转化过程中,AOB和anammox菌协作共生,AOR和NRR随
$ {\rm{N}}{{\rm{H}}^ +_4} $ -N质量浓度的提升稳步增加,分别达0.98 kg·(m3·d)−1和1.60 kg·(m3·d)−1左右,且anammox和亚硝化途径对$ {\rm{N}}{{\rm{H}}^ +_4} $ -N的转化比稳定在0.73左右。3)由anammox转为CANON工艺过程中,anammox菌和AOB基因丰度最终分别达4.41×1010拷贝数·g−1和1.81×1011拷贝数·g−1。Anammox菌在CANON启动前期和高负荷条件下分别以Candidatus Kuenenia和Candidatus Brocadia为优势菌属,而SM1A02作为可能的anammox菌属同氨氧化菌属Nitrosomona在启动过程中始终为优势菌属,最终AOB和anammox菌分别占细菌总数的18.95%和13.17%。







 下载:
下载: