-
在短程硝化反硝化、同步硝化反硝化、厌氧氨氧化等[1]非传统生物脱氮工艺的生物脱氮系统中,氨化菌会在好氧条件下将有机氮转化为氨氮(
${\rm{NH}}_4^{+} $ -N),然后氨氧化菌(ammonia-oxidizing bacteria,AOB)和亚硝酸盐氧化菌(nitrite-oxidizing bacteria,NOB)会协同作用将氨氮(${\rm{NH}}_4^{+} $ -N)转化为硝态氮(${\rm{NO}}_3^{-} $ -N)。该协同作用的过程即AOB先将${\rm{NH}}_4^{+} $ -N氧化为亚硝态氮(${\rm{NO}}_2^{-} $ -N),NOB再将${\rm{NO}}_2^{-} $ -N进一步氧化为${\rm{NO}}_3^{-} $ -N[2]。其中,NOB为革兰氏阴性菌,是自养微生物,其适宜的生长温度为25~30 ℃[3]。在好氧条件下,NOB以CO2为碳源参与硝化反应,以氧气和硝酸盐为电子受体进行亚硝酸盐的氧化。此外,NOB还可利用其他能源参与微生物的代谢活动,如NOB作为专性氧化菌以甲酸、硫化物或其他有机化合物为底物发生代谢反应[4]。根据微生物的形态特征和细胞内膜的排列方式,NOB可划分为7个属:硝化杆菌属(Nitrobacter)、硝化球菌属(Nitrococcus)、硝化刺菌属(Nitrospina)、硝化螺菌属(Nitrospira)、Nitrotoga属、Candidatus Nitromaritima属和Nitrolancea属[5]。其中,Nitrobacter和Nitrospira是污水生物处理系统中的主导NOB[6]。在高溶解氧、高底物浓度条件下,Nitrobacter含量远高于Nitrospira,而在低溶解氧和低基质底物浓度条件下,Nitrospira为优势菌属[7-8]。温度是硝化反应过程重要的影响因素之一。以溶解氧(dissolved oxygen,DO)和总氨氮(total ammonia nitrogen,TAN)为限制条件,当温度每升高1℃(在20 ℃基础上)时,硝化速率可分别提高1.108%和4.275%[9]。陈翠忠等[10]采用缺氧-好氧SBR处理合成废水时发现,当温度从15 ℃升至25 ℃时,氨氮去除率随之升高;当温度从25 ℃升至35 ℃时,氨氮去除率却逐渐降低。TAYLOR等[11]在测定
${\rm{NO}}_2^{-} $ 氧化动力学参数时发现,反应温度为30 ℃时的最大反应速率(Vmax)大于17 ℃时的Vmax。于雪等[12-13]发现:当温度较低时,微生物细胞膜呈凝胶状,水分和营养物质跨膜运输受阻,此时的细胞因缺乏营养而停止生命活动,反应速率随之降低;随着温度的升高,生化反应速率逐渐恢复并加快,且当温度超过一定范围后,细菌体内的蛋白质和核酸等物质对温度敏感的细胞组分发生变性,并使得其生长停止、最终死亡。近年来,16S rRNA基因Illumina MiSeq高通量测序技术被广泛应用于海洋、湖泊、水库、污水处理系统等水体微生物多样性、群落结构及功能研究领域[14-15]。众多学者基于该技术研究了温度对硝化菌群组成的影响,发现温度会影响NOB菌群的多样性和结构[16-19],但鲜有研究涉及NOB菌群的微生物代谢功能及相关功能基因。
本研究基于16S rRNA高通量测序技术,探究在3种温度(25 ℃、35 ℃和40 ℃)条件下,人工模拟废水中富集的NOB菌群间的关联网络关系、生物特征标记物及代谢功能,并利用KEGG数据库分析硝化过程微生物代谢途径,以期进一步揭示温度对硝化过程中的微生物的影响,为生物脱氮工艺的优化调控提供参考。
-
在实验采用的人工模拟废水中,
${\rm{NO}}_2^{-} $ -N的质量浓度通过投加NaNO2来调节(5.0~140 mg·L−1);初始pH利用浓度为1 mol·L−1的盐酸溶液和浓度为1 mol·L−1的氢氧化钠溶液来调节。为满足微生物自身增殖的需要,向每升模拟废水中添加的微量元素为:0.4 g NaHCO3;1 g KH2PO4;1.31 g K2HPO4;1.25 g EDTA;0.55 g ZnSO4·7H2O;0.40 g CoCl2·6H2O;1.275 g MnCl2·4H2O;0.40 g CuSO4·5H2O;0.05 g Na2MoO4·2H2O;1.375 g CaCl2·2H2O;1.25 g FeCl3·6H2O;44.4 g MgSO4·7HO。实验接种污泥取自兰州市西固区兰炼污水处理厂。该污泥具有良好的生物脱氮性能,其混合液挥发性悬浮固体浓度(MLVSS)为3 300 mg·L−1。
实验装置为SBR(有效容积为3 L,排水比为1∶3)。反应器的进出水
${\rm{NO}}_2^{-} $ -N质量浓度及MLVSS采用标准方法测定[20-21]。采用WTW Multi-3420多参数测定仪对温度、pH及DO进行实时监测。 -
在实验开始前对接种污泥进行了为期30 d的培养驯化,以获得稳定的硝化性能。在SBR内,不同阶段泥水混合液的温度分别控制为25 ℃(阶段Ⅰ)、35 ℃(阶段Ⅱ)和40 ℃(阶段Ⅲ)。每个阶段SBR分别运行100个周期,以获得相应温度条件下成熟的活性污泥。SBR的各运行周期由持续进水(1 min)、硝化(通过实时监测pH和DO控制硝化终点,硝化的同时曝气)、沉淀(30 min)、排水(5 min)和闲置5个阶段组成。此外,为避免pH和DO成为亚硝酸盐氧化的限制因素,pH维持在(7.5±0.1),DO维持在4.0~4.5 mg·L−1。利用比亚硝态氮氧化速率(SNiOR)来反映NOB的活性,其计算公式为式(1)。
式中:SNiOR为比亚硝态氮氧化速率,g·(g·d)−1(以每日每克可挥发性悬浮物氧化的亚硝态氮的质量计);c0(
${\rm{NO}}_2^{-} $ -N)为曝气开始时${\rm{NO}}_2^{-} $ -N的质量浓度,mg·L−1;ct(${\rm{NO}}_2^{-} $ -N)为曝气结束时${\rm{NO}}_2^{-} $ -N的质量浓度,mg·L−1;MLVSS为污泥的质量浓度,mg·L−1;tN为曝气时间,min[22]。 -
在25 ℃(第99周期)、35 ℃(第199周期)和40 ℃(第299周期)时,取SBR中的活性污泥样品(每个温度包括3个平行样品)并提取其DNA。将样品中提取到的DNA,对16S rRNA V3-V4区进行扩增,以活性污泥样本所提取到的DNA原液为PCR模板,通用引物序列为:F:520F(5-ACTCCTACGGGCAGCA-3)R:802R:(5-GGACTACHVGGGTWTCTAAT-3)。PCR反应条件为:98 ℃时预变性2 min;98 ℃时变性15 s;55 ℃时退火30 s;72 ℃时延伸30 s后再保温5 min,提取后于4 ℃下保存。委托上海派森诺生物科技有限公司进行Illumina MiSeq高通量测序,通过16S rRNA高通量进行测序分析并获得微生物的结构组成。在此基础上,利用KEGG数据库,借助PICRUSt工具预测分析微生物菌群的代谢功能,并对氮转化的相关功能基因进行比对。其中,相关性分析基于SPSS统计分析软件完成,LEfSe分析借助派森诺云分析平台完成。
-
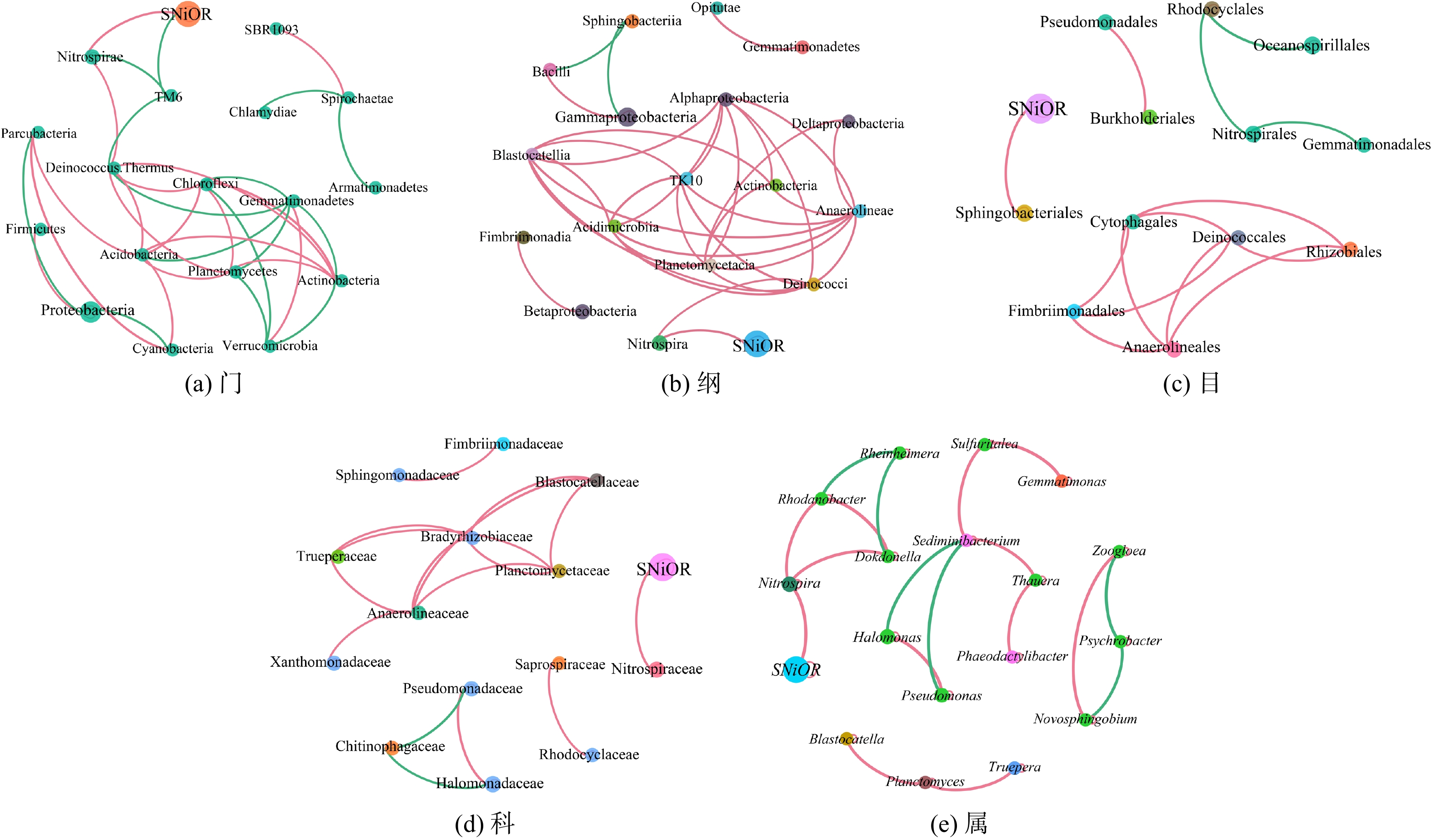
基于微生物的关联网络图可分析系统微生物群落之间、微生物与环境因子之间的相互关系。通过关联网络分析的方法,可探究系统内与微生物种群相互“协作”或“竞争”的模式。图1为不同温度条件下各分类水平(门phylum、纲class、目order、科family和属genus)的微生物种群与SNiOR的关联网络特征。图中相同颜色的节点为同一门,且节点越大,该物种在微生物类群中所占的相对丰度越大。而与节点相连的线越多,则表明该物种与其他微生物联系越密切。两节点间的连线表示微生物两两之间存在相关性(Spearman相关系数)。其中,红线表示正相关,绿线表示负相关。在各分类水平上,各微生物间具有错综复杂的交互关系,且呈正相关的微生物多于呈负相关的微生物。此外,在亚硝酸盐氧化过程中,基质底物为亚硝酸盐,故SNiOR是NOB活性的直接体现。关联网络图表明,SNiOR与硝化螺菌门Nitrospirae、硝化螺菌纲Nitrospirae、鞘脂杆菌目sphingobacterials、鞘脂杆菌科sphingobacterials及硝化螺菌属Nitrospira均表现出正相关性。Nitrospira是污水生化处理系统中NOB菌群的代表菌属[23-25]。在本研究中,SNiOR与Nitrospira呈现正相关关系(Spearman相关系数ρ为0.95)。此外,对于生化系统内的Nitrospira,与其呈正相关的分别为黑罗河杆菌属(Rhodanobacter)(ρ=0.88)和独岛菌属(Dokdonella)(ρ=0.88),而未发现与Nitrospira呈负相关关系的微生物菌属。在各个分类水平上,SNiOR仅与TM6门呈负相关关系(ρ=−0.87)。这表明,在生化系统中亚硝酸盐的氧化是基于多种微生物协同作用来完成的,且较多的微生物种群间以相互“协作”作用共存,仅较少的微生物种群间存在相互“竞争”的生化反应。
-
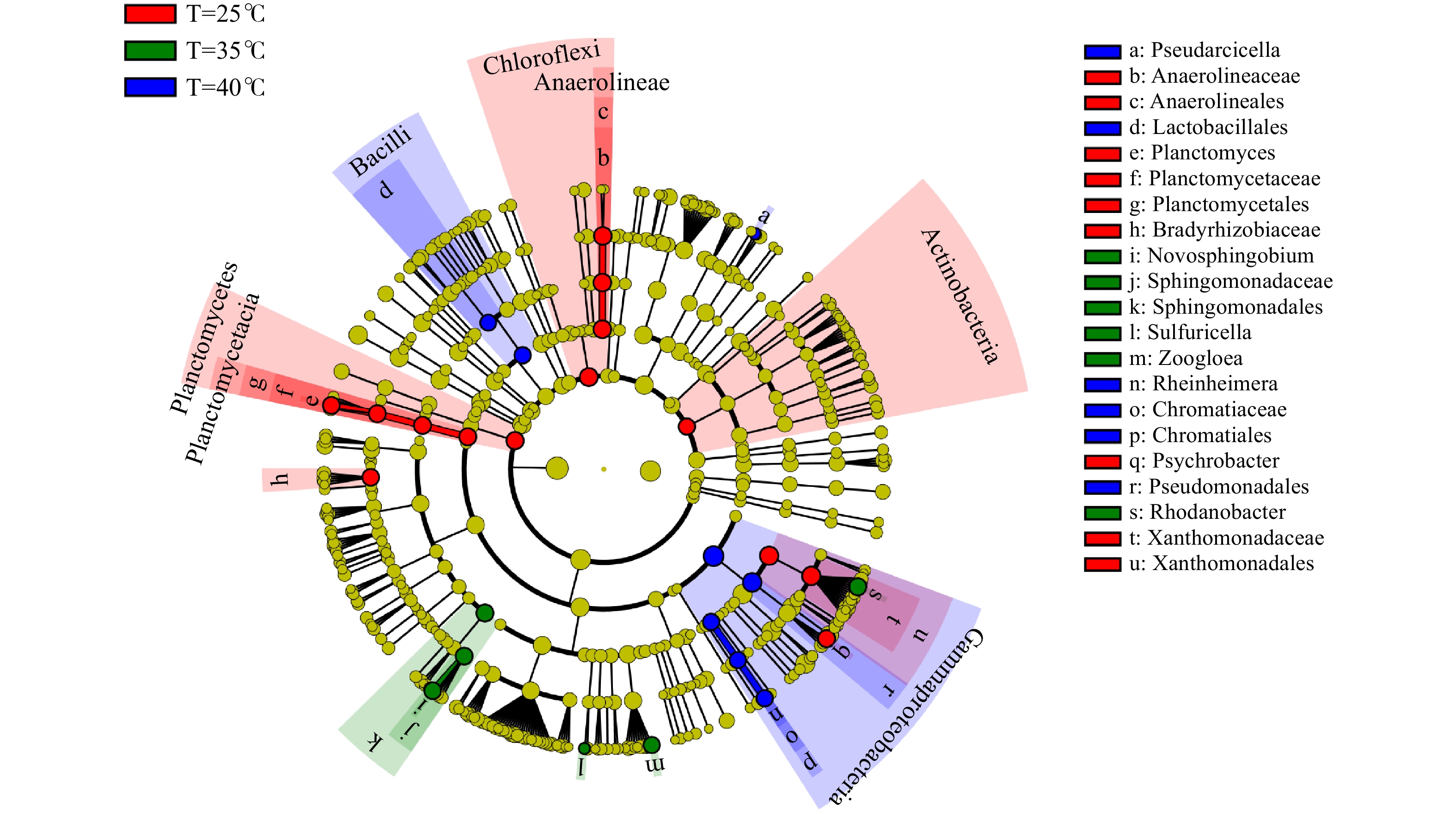
LefSe是一种基于线性判别分析(linear discriminant analysis,LDA)效应量的分析方法,其本质是将线性判别分析与非参数的Kruskal-Wallis及Wilcoxon秩和检验相结合,从而筛选关键生物标记物(即关键群落成员)。在3种温度条件下,采用LefSe筛选出不同温度条件下污泥样品的关键生物,结果如图2所示。活性污泥样品的分类等级树展示了微生物群落中从门到属(从内圈到外圈依次排列)所有微生物的等级关系。其中,节点大小对应于该物种的平均相对丰度,黄色圆点代表未体现出显著组间差异的微生物物种,而其他颜色(如绿色和红色)则表明这些物种呈现显著组间差异,且在该色所代表分组样品中丰度较高。
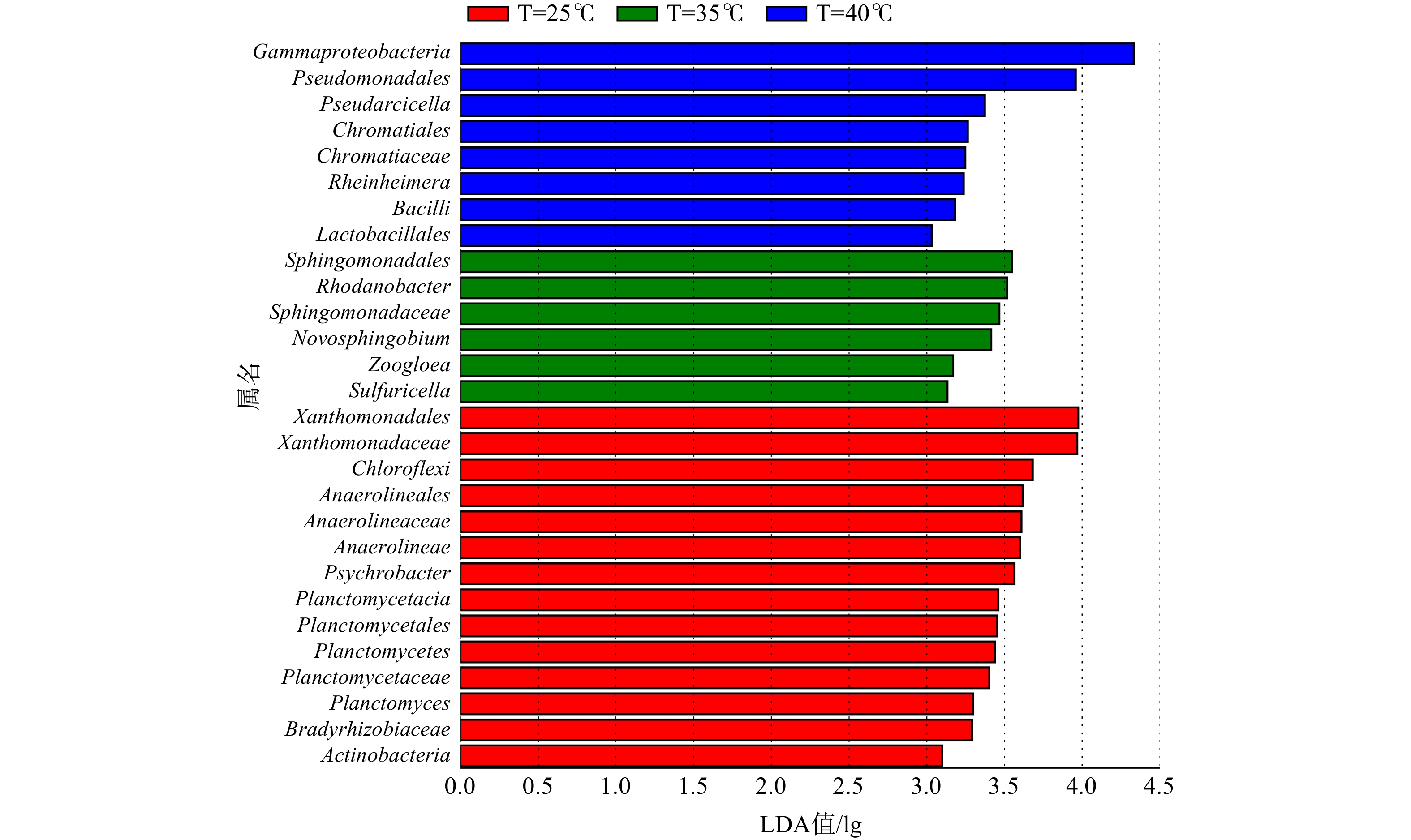
此外,通过线性判别分析(LDA值)方法进行了生物标记分析,以获得不同温度样品间具有显著差异的菌群。图3为在3种温度条件下污泥样品组间LDA值大于3.0的具有显著差异的微生物种群,柱状图的长度代表差异物种的显著性(即为LDA值)。在25℃、35℃和40℃温度条件下,共筛选出28种具有显著差异的微生物标记物。每种温度条件下的生物标记物分别有14种、6种和8种。以35℃活性污泥样品为例,SBR生化系统中存在Sphingomonadales(4.1%)、Rhodanobacter(4.3%)、Sphingomonadaceae(3.5%)、Novosphingobium(2.7%)、Zoogloea(1.8%)、Sulfuricella(0.02%)等6种生物标记物。在该温度下,这些微生物在系统内具有相对较高的丰度,是微生物种群的优势菌属。
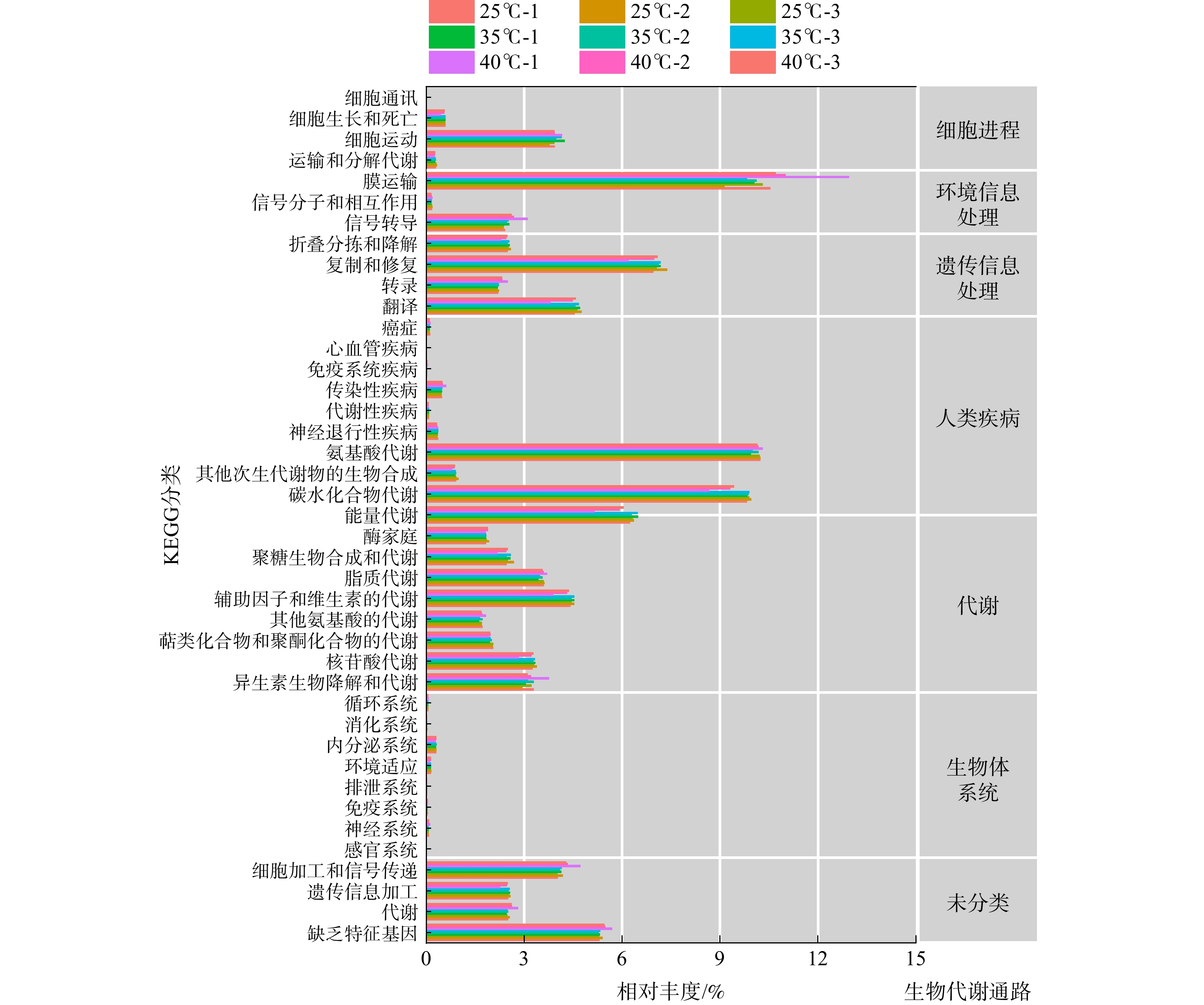
-
将上述研究得到的基因组数据与KEGG数据库进行了比对,以推测系统中微生物代谢通路并分析其代谢功能。在分类水平上,KEGG数据库包括4个等级:第一等级为生物代谢通路;第二等级为代谢通路的子功能;第三等级为代谢通路图;第四等级为KO 编号(对应代谢通路上各个KO的具体注释信息)[26]。图4为基于KEGG数据库的3种温度条件下活性污泥微生物代谢功能分析结果。通过数据比对获得了7类生物代谢通路功能,即细胞进程、环境信息处理、遗传信息处理、人类疾病、新陈代谢、生物体系统和未分类这7个功能模块。在3种温度下,KEGG一级代谢通路中7个功能模块丰度变化趋势一致。即代谢功能模块在KEGG一级代谢通路中基因丰度最高,环境信息处理、遗传信息处理以及未分类的功能模块相应的基因丰度次之,生物体系统和人类疾病模块相关基因丰度较低。在代谢功能模块中,氨基酸代谢、碳水化合物代谢和能量代谢是最大的功能类群,这三者分别占总生物代谢通路的10.2%、9.9%和6.3%(在3种温度下取平均值,以下同理);其次是辅助因子和维生素的代谢、脂质代谢和核苷酸代谢等;环境信息处理模块中膜运输占总生物代谢通路的10.5%,信号转导占2.4%;遗传信息处理模块中,对遗传信息的复制和修复、翻译占据主要优势,其相对丰度分别为7.1%和4.6%;未分类功能模块中,细胞加工和信号传递、遗传信息处理以及代谢功能分别占总生物代谢通路的4.0%、2.5%和2.5%。
以上结果表明,低温有利于微生物对碳水化合物的代谢、能量代谢;较高温度则会抑制能量代谢,即影响微生物的新陈代谢。因此,应避免在高温下运行以硝化反应占主导的脱氮工艺系统,以避免降低其中微生物的活性。
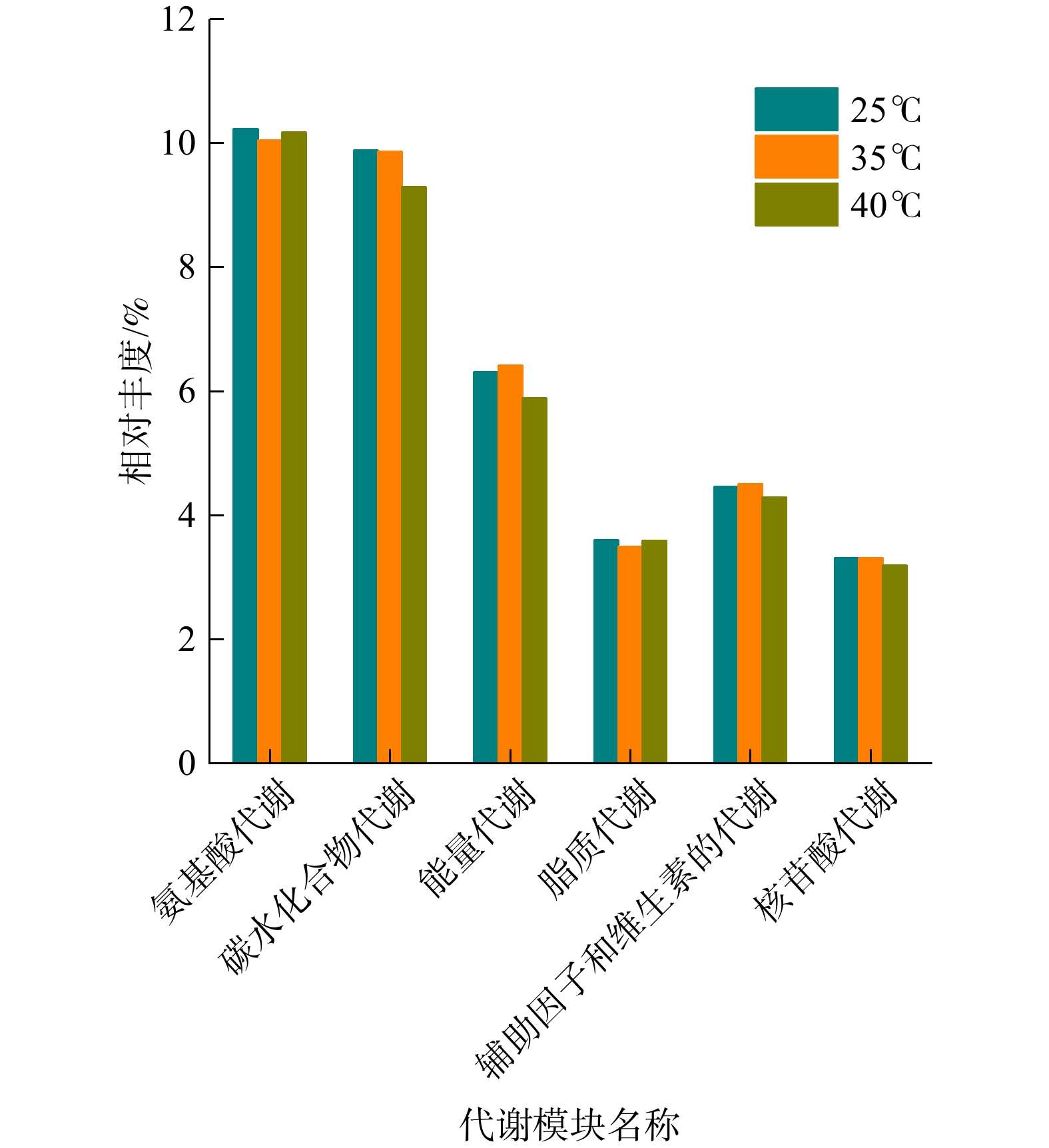
为进一步研究温度对微生物在脱氮工艺中代谢功能的影响,本研究建立了KEGG二级代谢模型(图5),即在3种温度下6种代谢功能的变化规律。随着温度的升高(25 ℃→35 ℃→40 ℃),系统中微生物氨基酸代谢、辅助因子和维生素的代谢、脂质代谢和核苷酸代谢的相对丰度变化较小,而对碳水化合物及能量的代谢在35 ℃升至40 ℃的过程中呈下降趋势。此外,NOB氧化亚硝酸盐的过程属于能量代谢,且随着温度升高,能量代谢的相对丰度表现为先升高、后降低的趋势。
在3种温度条件下,一级代谢通路中7个功能模块丰度变化趋势一致,代谢功能模块基因丰度最高。但在不同温度条件下,KEGG二级代谢相对丰度发生变化。NOB 降解
${\rm{NO}}_2^{-} $ -N的代谢功能随着温度的升高,呈现先升高、后降低的趋势。KO一级水平层次上的7类代谢功能和KO二级水平层次上的 41 类二级代谢功能均具有显著的丰度差异。这种丰度差异在编码新陈代谢的基因序列模块中体现尤为突出,表明在活性污泥样品中存在大量具有特定生态功能的微生物菌群,其新陈代谢活动较为活跃。 -
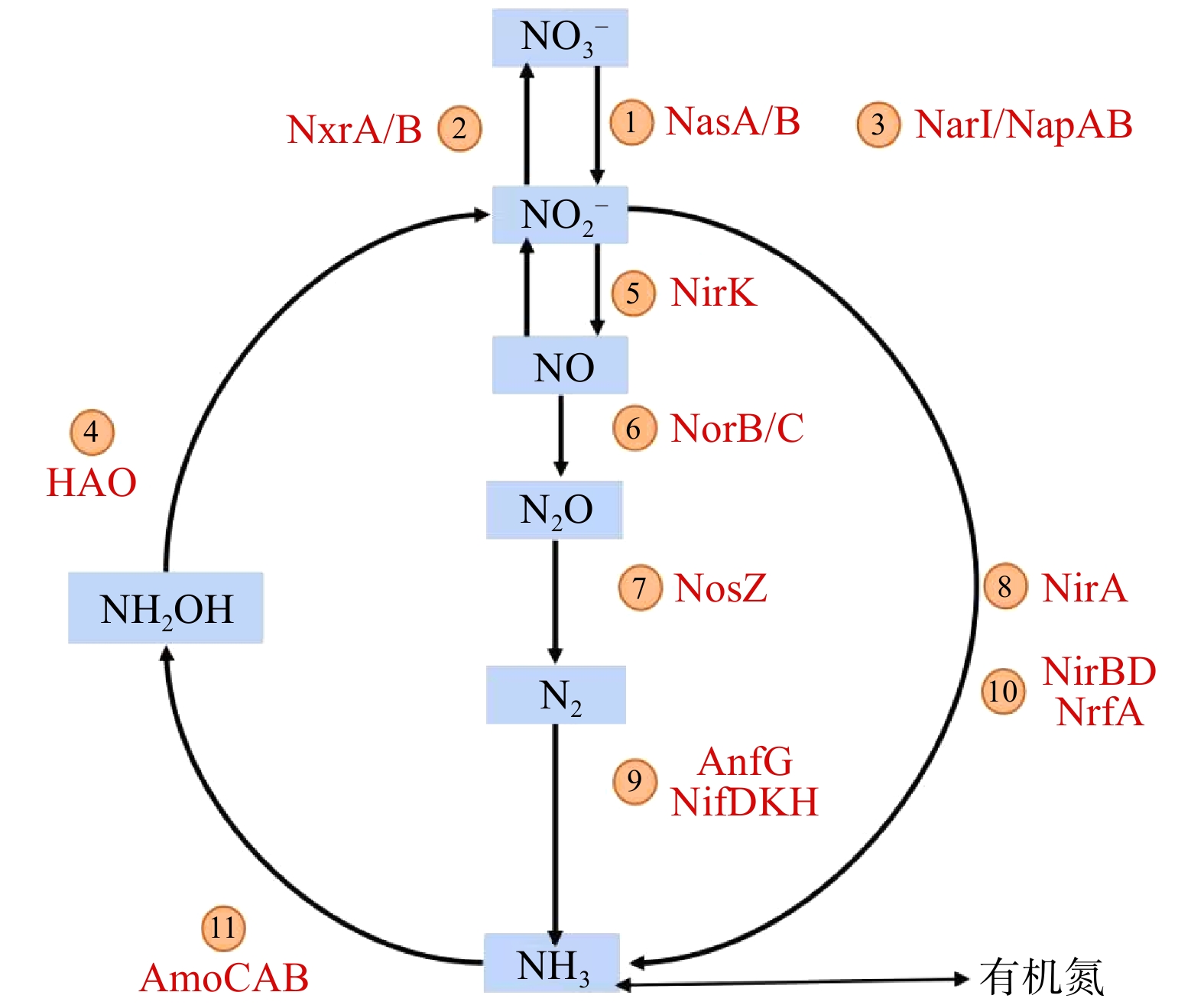
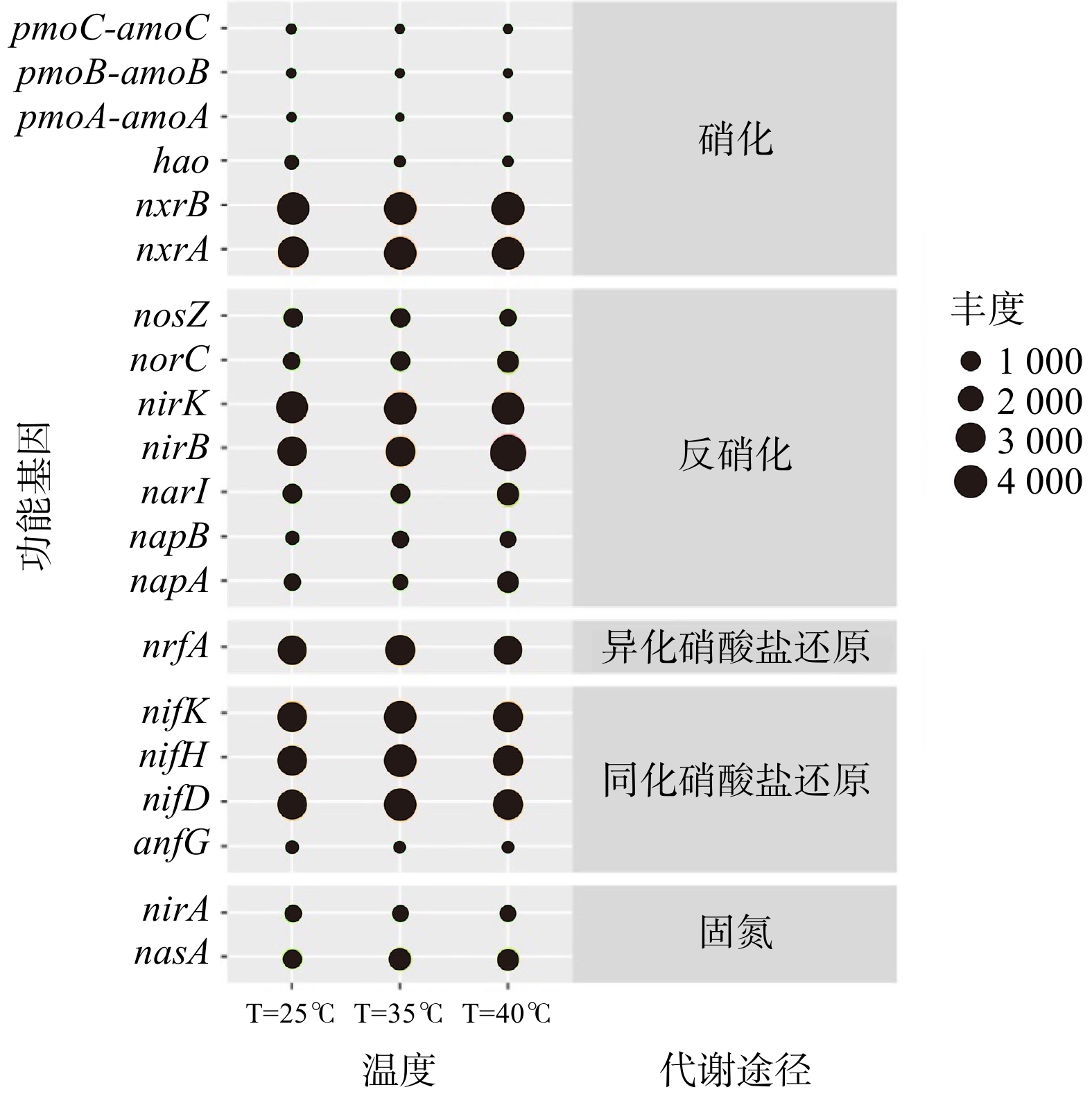
基于16S rRNA基因测序结果和PICRUSt工具可获得KEGG直系同源基因簇(KO)的具体注释信息。将注释得到的所有KO值映射到KEGG代谢通路数据库的“氮代谢通路图”中,可比对出与氮代谢相关的功能基因及其相对丰度。温度批次实验的所有样品均存在5种氮代谢途径(硝化、异化硝酸盐还原、同化硝酸盐还原、反硝化和固氮)和24个功能基因(图6)。硝化过程相关的功能酶为氨单加氧酶(amoABC)和羟胺氧化酶(hao)/还原酶(hcp)及亚硝酸盐氧化还原酶(nxrA、nxrB)。在3种温度下,亚硝酸盐氧化还原酶(nxrA、nxrB)丰度(9.9%、10.5%、10.3%)均远远高于氨单加氧酶(AmoCAB)和羟胺脱氢酶(hao)丰度总和(0.3%、0.3%、0.1%)。这表明亚硝酸盐氧化作用在系统硝化过程中占据主导地位。
对比在3种温度条件下氮转化功能基因的丰度可发现,相关酶存在于活性污泥样品的氮转化过程中,但其功能基因的相对丰度存在一定差异。5种氮转化过程对应的功能基因如图7所示。在25 ℃、35 ℃和40 ℃条件下,亚硝酸盐氧化还原酶(nxr)类基因(nxrA和nxrB)相对丰度分别为9.9%、10.5%和10.3%,随着温度的升高,基因数量基本保持不变。此外,在硝化过程中存在催化氨为羟胺的氨单加氧酶类基因(AmoABC)、将羟胺转化为亚硝酸盐的羟胺脱氢酶(hao),以及将亚硝酸盐氧化为硝酸盐的亚硝酸盐氧化还原酶(nxrA、nxrB),其中nxrA、nxrB分别为NOB菌属中硝化杆菌属(Nitrobactor)和硝化螺菌属(Nitrospira)的功能基因。这2种菌属为硝化过程中的关键菌属,可将
${\rm{NO}}_2^{-} $ -N氧化为${\rm{NO}}_3^{-} $ -N。硝化过程参与的酶相对丰度大小为:nxrB>nxrA>hao>AmoCAB。这表明系统中存在大量将${\rm{NO}}_2^{-} $ -N氧化为${\rm{NO}}_3^{-} $ -N的基因,亦说明整个系统内的亚硝酸盐氧化效果显著。 -
1)在3种温度条件下,多种生物的协同作用是实现亚硝酸盐氧化的生物反应机制。SNiOR仅与Nitrospira呈现正相关性(Spearman系数ρ=0.95),而Nitrospira相对丰度是SNiOR的唯一影响因子。通过LefSe分析表明,微生物种群的群落结构也存在显著差异,分别获得了14种、6种和8种具有显著差异(P<0.05)的微生物标记物。
2)温度对微生物的代谢功能有一定程度的影响。碳水化合物代谢、能量代谢、氨基酸代谢、核苷酸代谢及酶类代谢共存于活性污泥系统中,而能量代谢、核苷酸代谢和酶类代谢是微生物新陈代谢的主导功能模块,且低温有利于碳水化合物代谢、能量代谢。较高温度会抑制能量代谢,NOB丰度降低。
3)温度对氮代谢有关基因丰度的影响并不显著。随着温度的升高,亚硝酸盐氧化还原酶(nxr)类基因(nxrA和nxrB)的数量基本保持不变。此外,对硝化过程的基因统计结果表明,具有亚硝酸盐氧化功能的基因相对丰度远大于具有氨氧化作用的基因相对丰度,系统的亚硝酸盐氧化效果显著。
利用16S rRNA高通量测序技术考察温度对生物脱氮硝化过程中亚硝酸盐氧化菌代谢功能的影响
Investigation on the effect of temperature on the metabolism and function of nitrite-oxidizing bacteria in the process of biological nitrification by 16S rRNA high-throughput sequencing technology
-
摘要: 为探究温度对生物脱氮硝化过程微生物代谢及功能基因的影响,采用16S rRNA高通量测序技术,以富集的亚硝酸盐氧化菌(NOB)为研究对象,考察了活性污泥系统硝化过程中菌属与比亚硝酸盐氧化速率(SNiOR)的相关性。通过LEfSe分析筛选出3种温度(25、35和40℃)条件下的生物标记物,并利用KEGG数据库解析了微生物的代谢功能、氮代谢相关酶类型及相对丰度。结果表明,在3种温度条件下,NOB富集系统内的Nitrospira是SNiOR的唯一影响因子,且两者呈现显著的正相关性,Spearman相关系数(ρ)为0.95。此外,在温度条件为25、35和40℃时,分别筛选出14种、6种和8种生物标记物,显著优势菌分别是Xanthomonadales目、Sphingomonadales目和Gammaproteobacteria纲。以上结果说明:温度对微生物的代谢功能具有一定程度的影响,低温有利于碳水化合物代谢、能量代谢;较高温度会抑制能量代谢,NOB丰度降低;随着温度的升高,亚硝酸盐氧化还原酶(nxr)类基因(nxrA和nxrB)的数量基本保持不变。Abstract: In order to investigate the effect of temperature on microbial metabolism and functional genes during biological nitrification, 16S rRNA high-throughput sequencing technology was used to investigate the correlation between bacterial genera and specific nitrite oxidation rate (SNiOR) in the nitrite oxidizing bacteria (NOB) enrichment activated sludge system. The biomarkers were screened by LEfSe analysis at three different temperatures (25°C, 35°C and 40°C). Based on the KEGG database, the metabolic functions, nitrogen metabolism-related enzyme types and relative abundance of microorganisms were also resolved. The results showed that Nitrospira was the only influencing factor of SNiOR within the NOB enrichment system at three different temperatures , and there was a significant positive correlation between Nitrospira and SNiOR with the Spearman correlation coefficient (ρ) at 0.95. In addition, 14, 6 and 8 biomarkers were screened at 25°C, 35°C and 40°C, respectively. The dominant genera were Xanthomonadales, Sphingomonadales and Gammaproteobacteria, respectively. These results suggested that temperature had a certain degree of influence on the metabolic function of microorganisms. Low temperature favored carbohydrate metabolism and energy metabolism, and higher temperature inhibited energy metabolism and decreased NOB abundance. The number of nitrite oxidoreductase (nxr) genes (nxrA and nxrB) remained almost unchanged with the increase of temperature.
-

-
-
[1] 马智明. 生物脱氮除磷理论与技术进展[J]. 化工管理, 2018(21): 179-180. doi: 10.3969/j.issn.1008-4800.2018.21.137 [2] PENG Y Z, ZHU G B, GUO J H, et al. Biological nitrogen removal with nitrification and denitrification via nitrite pathway[J]. Applied Microbiology and Biotechnology, 2006, 73(1): 15-26. doi: 10.1007/s00253-006-0534-z [3] WANG T, ZHANG L G, LE B, et al. Simultaneous heterotrophic nitrification and aerobic denitrification at high concentrations of NaCl by Halomonas Bacteria[J]. IOP Conference Series:Earth and Environmental Science, 2019, 237: 052033. doi: 10.1088/1755-1315/237/5/052033 [4] 史银银, 明红霞, 陈泉睿, 等. 亚硝酸盐氧化细菌的生态位以及对海洋环境变化的响应机制[J]. 微生物学通报. 2021, 48(8) : 2985-2901. [5] DAIMS H, LÜCKER S, WAGNER M, et al. A new perspective on microbes formerly known as nitrite-oxidizing bacteria[J]. Trends in Microbiology, 2016, 24(9): 699-712. doi: 10.1016/j.tim.2016.05.004 [6] GOUDAR C T, GANJI S H, PUJAR B G. Substrate inhibition kinetics of phenol biodegradation[J]. Water Environment Research, 2000, 72(1): 50-55. doi: 10.2175/106143000X137103 [7] SPIECK E, HARTWIG C, CORMACK I, et al. Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge[J]. Environmental Microbiology, 2006, 8: 405-415. doi: 10.1111/j.1462-2920.2005.00905.x [8] 包鹏, 王淑莹, 马斌, 等. 不同溶解氧间歇曝气对亚硝酸盐氧化菌的影响[J]. 中国环境科学, 2016, 36(9): 2696-2702. doi: 10.3969/j.issn.1000-6923.2016.09.024 [9] ZHU S M, CHEN S. The impact of temperature on nitrification rate in fixed film biofilters[J]. Aquacultural Engineering, 2002, 26(4): 221-237. doi: 10.1016/S0144-8609(02)00022-5 [10] 陈翠忠, 高宇学, 王文迪, 等. 温度对SBR生物脱氮效能的长期影响[J]. 环境工程, 2018, 36(6): 68-72. [11] TAYLOR A E, MYROLD D D, BOTTOMLEY P J. Temperature affects the kinetics of nitrite oxidation and nitrification coupling in four agricultural soils[J]. Soil Biology and Biochemistry, 2019, 136: 107523-107523. doi: 10.1016/j.soilbio.2019.107523 [12] 于雪, 孙洪伟, 李维维, 等. 温度对硝化杆菌(Nitrobacter)活性动力学影响[J]. 中国环境科学, 2019, 40(3): 1427-1430. [13] 吴鹏, 陆爽君, 徐乐中, 等. 温度对ABR-MBR复合工艺处理生活污水的影响及其微生物群落分析[J]. 环境科学, 2014, 35(9): 3466-3472. [14] WERTZ S, POLY F, LE ROUX X, et al. Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil[J]. FEMS Microbiology Ecology, 2008, 63(2): 261-271. doi: 10.1111/j.1574-6941.2007.00416.x [15] CHU Y J, COREY D R. RNA sequencing: Platform selection, experimental design, and data interpretation[J]. Nucleic Acid Therapeutics, 2012, 22(4): 271-274. doi: 10.1089/nat.2012.0367 [16] 曾涛涛, 蒋小梅, 韩科昌, 等. 生活污水处理厂微生物群落结构解析[J]. 安全与环境学报, 2018, 18(2): 697-703. [17] FRIAS-LOPEZ J, SHI Y, TYSON G W, et al. Microbial community gene expression in ocean surface waters[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(10): 3805-3810. doi: 10.1073/pnas.0708897105 [18] 国家环境保护总局. 水和废水监测分析方法[M]. 4版. 北京: 中国环境科学出版社, 2002: 211-213 [19] HEWSON I, PORETSKY R S, TRIPP H J, et al. Spatial patterns and light-driven variation of microbial population gene expression in surface waters of the oligotrophic open ocean[J]. Environmental Microbiology, 2010, 12(7): 1940-1956. doi: 10.1111/j.1462-2920.2010.02198.x [20] 张婷婷, 张建, 杨芳, 等. 温度对污水脱氮系统污染物去除效果及氧化亚氮释放的影响[J]. 环境科学, 2012, 33(4): 1283-1287. [21] LANGILLE M G I, ZANEVELD J, CAPORASO J G, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences[J]. Nature Biotechnology, 2013, 31(9): 814-821. doi: 10.1038/nbt.2676 [22] WANG Q, YE L, JIANG G, et al. Side-stream sludge treatment using free nitrous acid selectively eliminates nitrite oxidizing bacteria and achieves the nitrite pathway[J]. Water Research, 2014, 55(10): 245-255. [23] MA B, YANG L, WANG Q, et al. Inactivation and adaptation of ammonia-oxidizing bacteria and nitrite-oxidizing bacteria when exposed to free nitrous acid[J]. Bioresource Technology, 2017, 245(8): 1266-1270. [24] RADAX R, RATTEI T, LANZEN A, et al. Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community[J]. Environmental Microbiology, 2012, 14(5): 1308-1324. doi: 10.1111/j.1462-2920.2012.02714.x [25] LI Y, LI J, WANG Z, et al. Inhibition kinetics of nitritation and half-nitritation of old landfill leachate in a membrane bioreactor[J]. Journal of Bioscience and Bioengineering, 2016, 123(4): 482-488. [26] 杨浩, 张国珍, 杨晓妮, 等. 典型集雨人饮地区窖水微生物群落多样性及差异解析[J]. 环境科学, 2017, 38(4): 4733-4746. -



 下载:
下载: